Alceu de S. Britto Jr.
Pattern Spotting and Image Retrieval in Historical Documents using Deep Hashing
Aug 04, 2022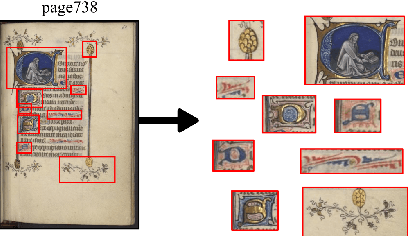
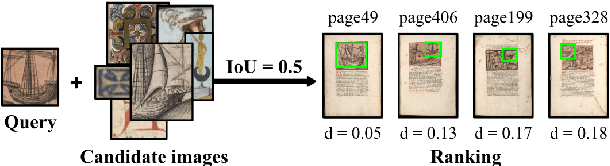
Abstract:This paper presents a deep learning approach for image retrieval and pattern spotting in digital collections of historical documents. First, a region proposal algorithm detects object candidates in the document page images. Next, deep learning models are used for feature extraction, considering two distinct variants, which provide either real-valued or binary code representations. Finally, candidate images are ranked by computing the feature similarity with a given input query. A robust experimental protocol evaluates the proposed approach considering each representation scheme (real-valued and binary code) on the DocExplore image database. The experimental results show that the proposed deep models compare favorably to the state-of-the-art image retrieval approaches for images of historical documents, outperforming other deep models by 2.56 percentage points using the same techniques for pattern spotting. Besides, the proposed approach also reduces the search time by up to 200x and the storage cost up to 6,000x when compared to related works based on real-valued representations.
Evaluation of Self-taught Learning-based Representations for Facial Emotion Recognition
Apr 26, 2022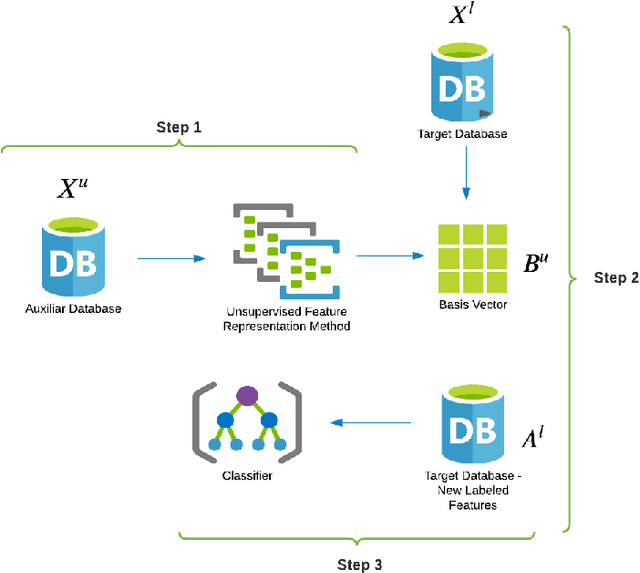
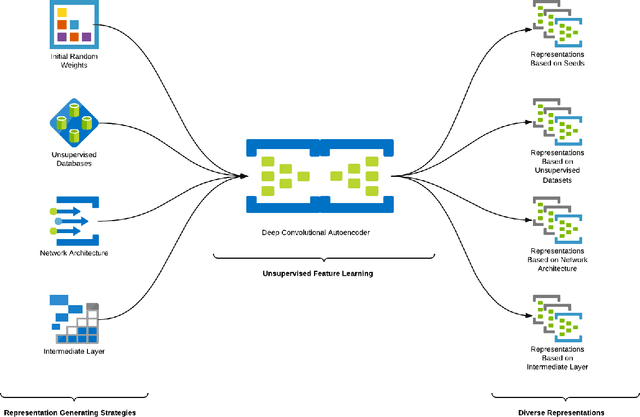


Abstract:This work describes different strategies to generate unsupervised representations obtained through the concept of self-taught learning for facial emotion recognition (FER). The idea is to create complementary representations promoting diversity by varying the autoencoders' initialization, architecture, and training data. SVM, Bagging, Random Forest, and a dynamic ensemble selection method are evaluated as final classification methods. Experimental results on Jaffe and Cohn-Kanade datasets using a leave-one-subject-out protocol show that FER methods based on the proposed diverse representations compare favorably against state-of-the-art approaches that also explore unsupervised feature learning.
Data Augmentation for Histopathological Images Based on Gaussian-Laplacian Pyramid Blending
Jan 31, 2020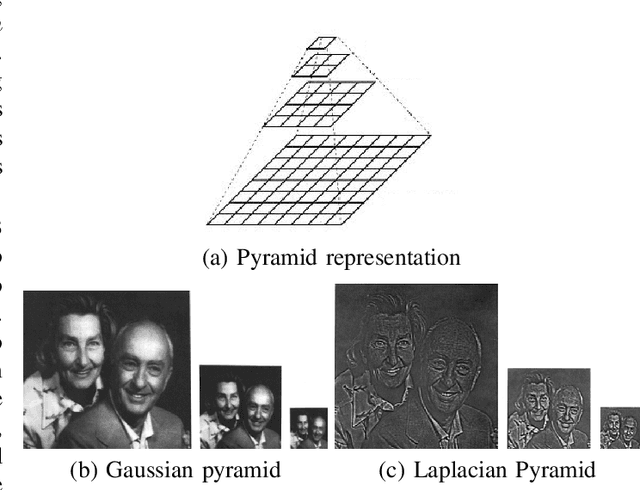
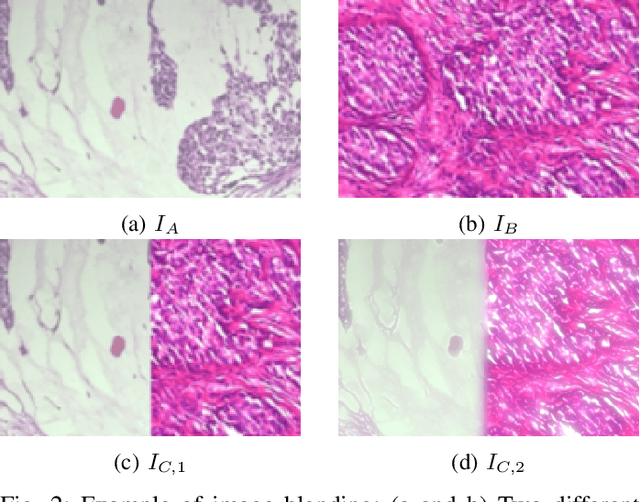
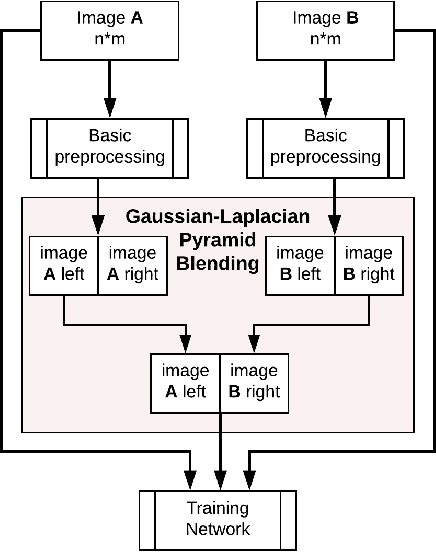
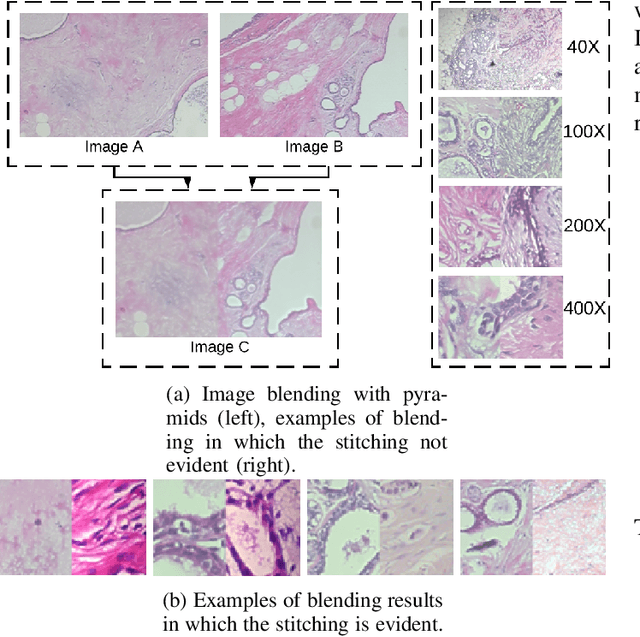
Abstract:Data imbalance is a major problem that affects several machine learning algorithms. Such problems are troublesome because most of the learning algorithms attempts to optimize a loss function based on error measures that do not take into account the data imbalance. Accordingly, the learning algorithm simply generates a trivial model that is biased toward predicting the most frequent class in the training data. Data augmentation techniques have been used to mitigate the data imbalance problem. However, in the case of histopathologic images (HIs), low-level as well as high-level data augmentation techniques still present performance issues when applied in the presence of inter-patient variability; whence the model tends to learn color representations, which are in fact related to the stain process. In this paper, we propose an approach capable of not only augmenting HIs database but also distributing the inter-patient variability by means of image blending using Gaussian-Laplacian pyramid. The proposed approach consists in finding the Gaussian pyramids of two images of different patients and finding the Laplacian pyramids thereof. Afterwards, the left half of one image and the right half of another are joined in each level of Laplacian pyramid, and from the joint pyramids, the original image is reconstructed. This composition, resulting from the blending process, combines stain variation of two patients, avoiding that color misleads the learning process. Experimental results on the BreakHis dataset have shown promising gains vis-\`a-vis the majority of traditional techniques presented in the literature.
Texture CNN for Histopathological Image Classification
May 28, 2019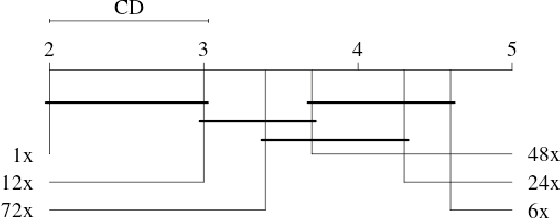
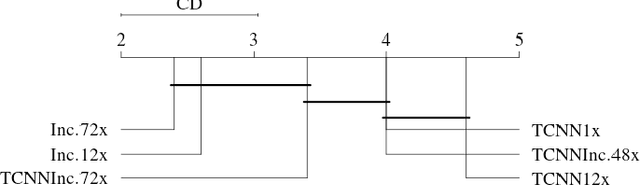
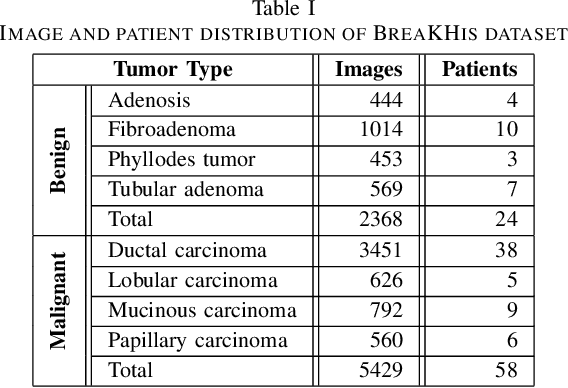
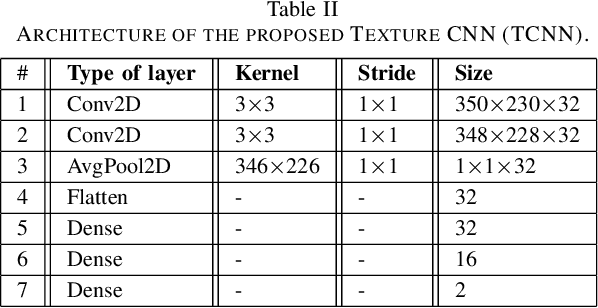
Abstract:Biopsies are the gold standard for breast cancer diagnosis. This task can be improved by the use of Computer Aided Diagnosis (CAD) systems, reducing the time of diagnosis and reducing the inter and intra-observer variability. The advances in computing have brought this type of system closer to reality. However, datasets of Histopathological Images (HI) from biopsies are quite small and unbalanced what makes difficult to use modern machine learning techniques such as deep learning. In this paper we propose a compact architecture based on texture filters that has fewer parameters than traditional deep models but is able to capture the difference between malignant and benign tissues with relative accuracy. The experimental results on the BreakHis dataset have show that the proposed texture CNN achieves almost 90% of accuracy for classifying benign and malignant tissues.
Double Transfer Learning for Breast Cancer Histopathologic Image Classification
Apr 16, 2019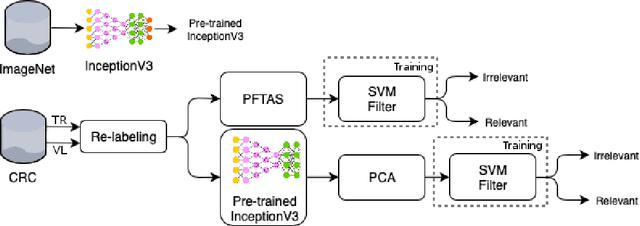
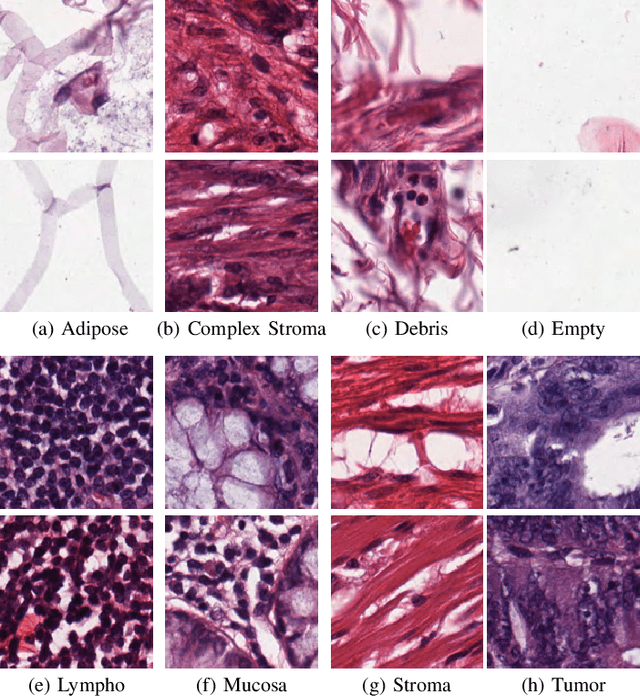
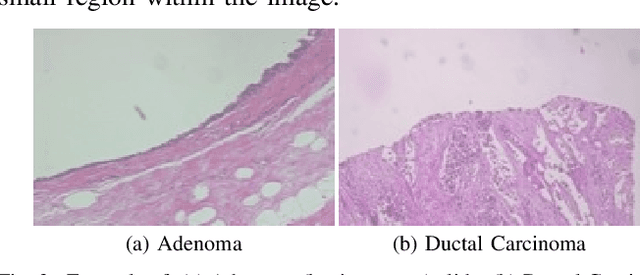
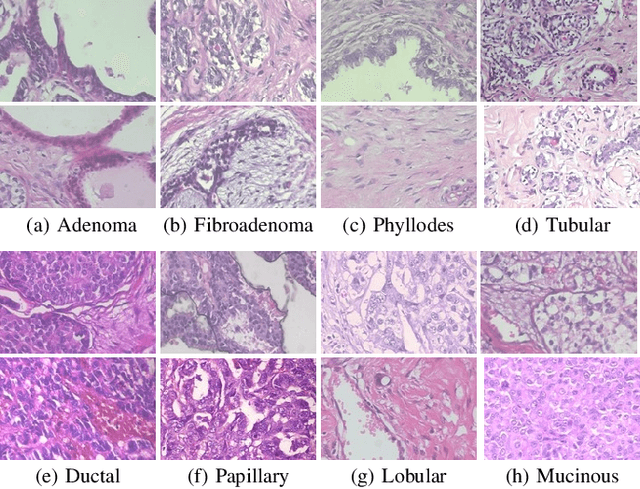
Abstract:This work proposes a classification approach for breast cancer histopathologic images (HI) that uses transfer learning to extract features from HI using an Inception-v3 CNN pre-trained with ImageNet dataset. We also use transfer learning on training a support vector machine (SVM) classifier on a tissue labeled colorectal cancer dataset aiming to filter the patches from a breast cancer HI and remove the irrelevant ones. We show that removing irrelevant patches before training a second SVM classifier, improves the accuracy for classifying malign and benign tumors on breast cancer images. We are able to improve the classification accuracy in 3.7% using the feature extraction transfer learning and an additional 0.7% using the irrelevant patch elimination. The proposed approach outperforms the state-of-the-art in three out of the four magnification factors of the breast cancer dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge