Steve Tsham Mpinda Ataky
Texture Characterization of Histopathologic Images Using Ecological Diversity Measures and Discrete Wavelet Transform
Feb 27, 2022
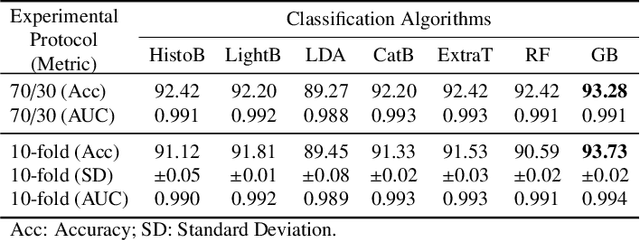
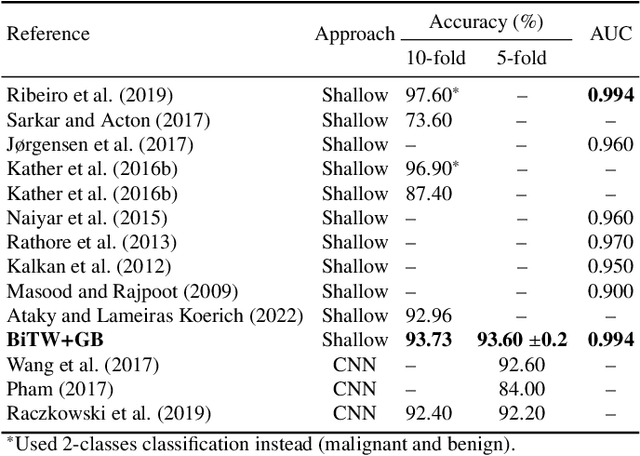
Abstract:Breast cancer is a health problem that affects mainly the female population. An early detection increases the chances of effective treatment, improving the prognosis of the disease. In this regard, computational tools have been proposed to assist the specialist in interpreting the breast digital image exam, providing features for detecting and diagnosing tumors and cancerous cells. Nonetheless, detecting tumors with a high sensitivity rate and reducing the false positives rate is still challenging. Texture descriptors have been quite popular in medical image analysis, particularly in histopathologic images (HI), due to the variability of both the texture found in such images and the tissue appearance due to irregularity in the staining process. Such variability may exist depending on differences in staining protocol such as fixation, inconsistency in the staining condition, and reagents, either between laboratories or in the same laboratory. Textural feature extraction for quantifying HI information in a discriminant way is challenging given the distribution of intrinsic properties of such images forms a non-deterministic complex system. This paper proposes a method for characterizing texture across HIs with a considerable success rate. By employing ecological diversity measures and discrete wavelet transform, it is possible to quantify the intrinsic properties of such images with promising accuracy on two HI datasets compared with state-of-the-art methods.
A Novel Bio-Inspired Texture Descriptor based on Biodiversity and Taxonomic Measures
Mar 07, 2021
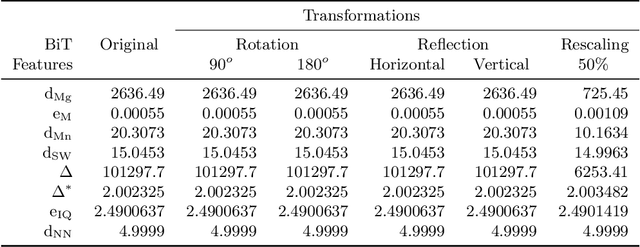
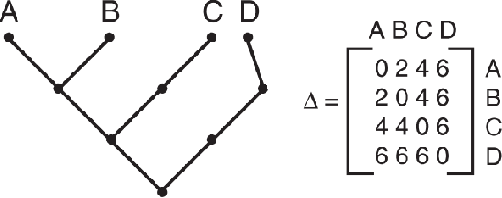
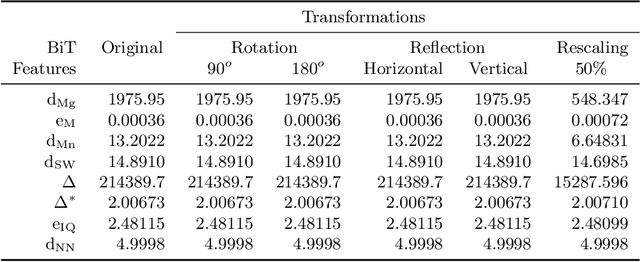
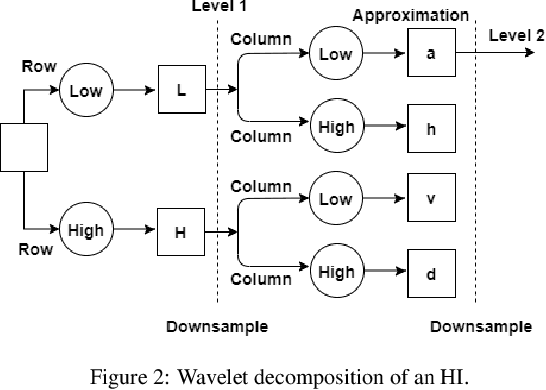
Abstract:Texture can be defined as the change of image intensity that forms repetitive patterns, resulting from physical properties of the object's roughness or differences in a reflection on the surface. Considering that texture forms a complex system of patterns in a non-deterministic way, biodiversity concepts can help texture characterization in images. This paper proposes a novel approach capable of quantifying such a complex system of diverse patterns through species diversity and richness and taxonomic distinctiveness. The proposed approach considers each image channel as a species ecosystem and computes species diversity and richness measures as well as taxonomic measures to describe the texture. The proposed approach takes advantage of ecological patterns' invariance characteristics to build a permutation, rotation, and translation invariant descriptor. Experimental results on three datasets of natural texture images and two datasets of histopathological images have shown that the proposed texture descriptor has advantages over several texture descriptors and deep methods.
Machine Learning Methods for Histopathological Image Analysis: A Review
Feb 07, 2021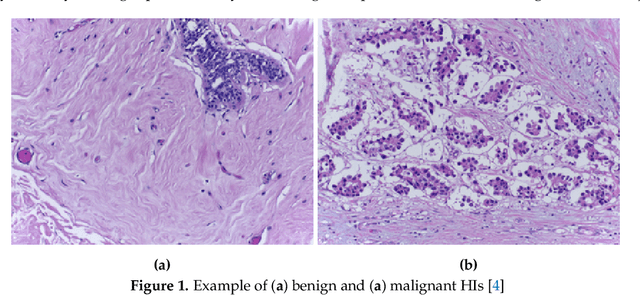
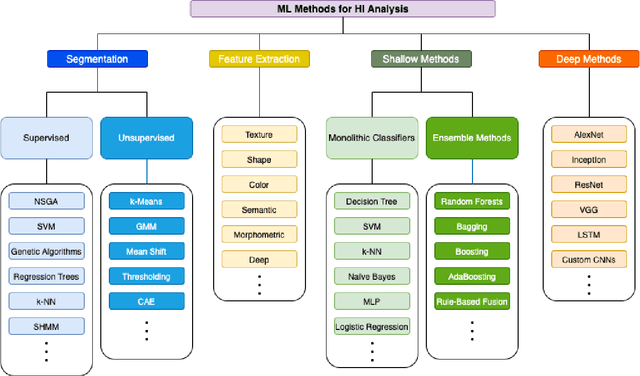
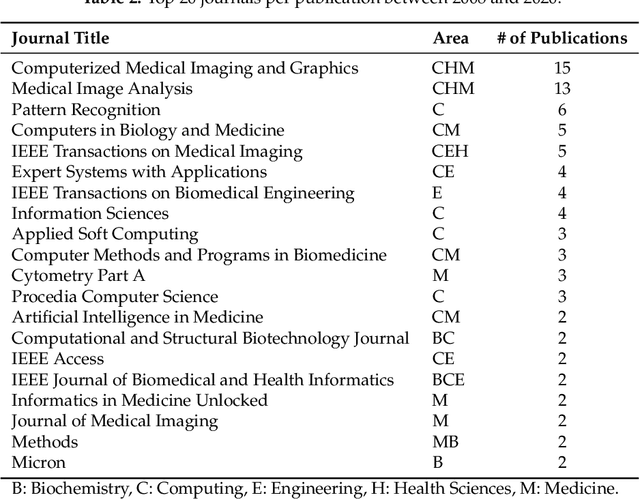
Abstract:Histopathological images (HIs) are the gold standard for evaluating some types of tumors for cancer diagnosis. The analysis of such images is not only time and resource consuming, but also very challenging even for experienced pathologists, resulting in inter- and intra-observer disagreements. One of the ways of accelerating such an analysis is to use computer-aided diagnosis (CAD) systems. In this paper, we present a review on machine learning methods for histopathological image analysis, including shallow and deep learning methods. We also cover the most common tasks in HI analysis, such as segmentation and feature extraction. In addition, we present a list of publicly available and private datasets that have been used in HI research.
Data Augmentation for Histopathological Images Based on Gaussian-Laplacian Pyramid Blending
Jan 31, 2020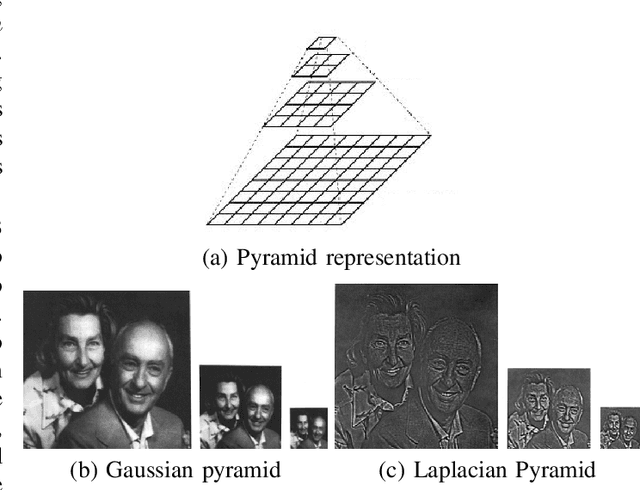
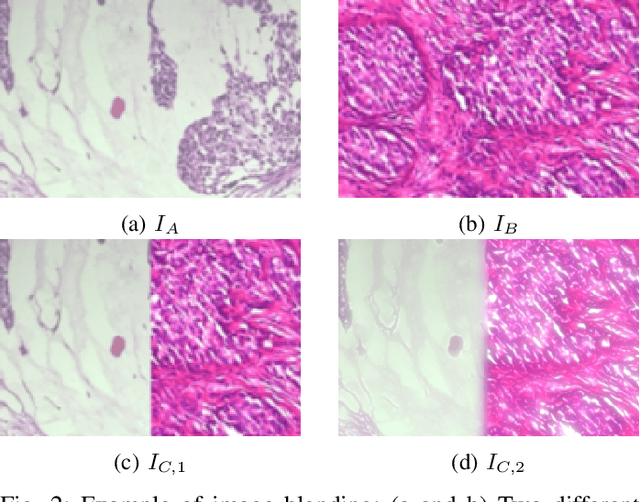
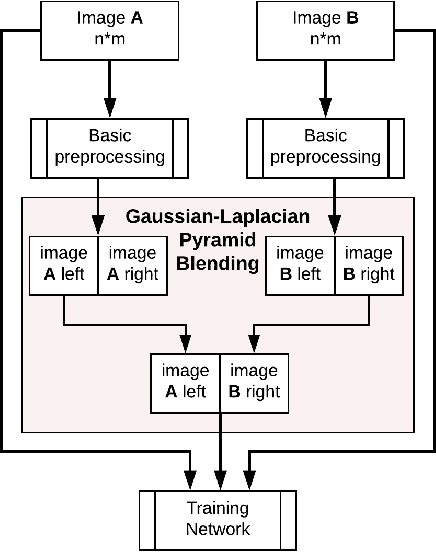
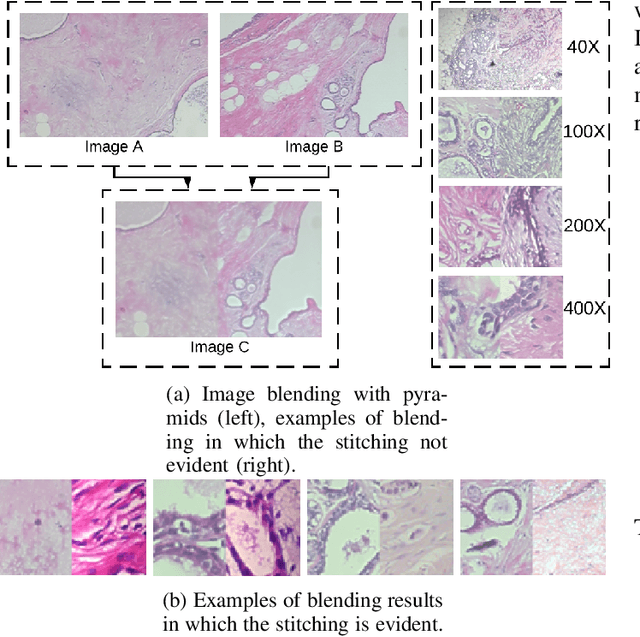
Abstract:Data imbalance is a major problem that affects several machine learning algorithms. Such problems are troublesome because most of the learning algorithms attempts to optimize a loss function based on error measures that do not take into account the data imbalance. Accordingly, the learning algorithm simply generates a trivial model that is biased toward predicting the most frequent class in the training data. Data augmentation techniques have been used to mitigate the data imbalance problem. However, in the case of histopathologic images (HIs), low-level as well as high-level data augmentation techniques still present performance issues when applied in the presence of inter-patient variability; whence the model tends to learn color representations, which are in fact related to the stain process. In this paper, we propose an approach capable of not only augmenting HIs database but also distributing the inter-patient variability by means of image blending using Gaussian-Laplacian pyramid. The proposed approach consists in finding the Gaussian pyramids of two images of different patients and finding the Laplacian pyramids thereof. Afterwards, the left half of one image and the right half of another are joined in each level of Laplacian pyramid, and from the joint pyramids, the original image is reconstructed. This composition, resulting from the blending process, combines stain variation of two patients, avoiding that color misleads the learning process. Experimental results on the BreakHis dataset have shown promising gains vis-\`a-vis the majority of traditional techniques presented in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge