Texture Characterization of Histopathologic Images Using Ecological Diversity Measures and Discrete Wavelet Transform
Paper and Code
Feb 27, 2022
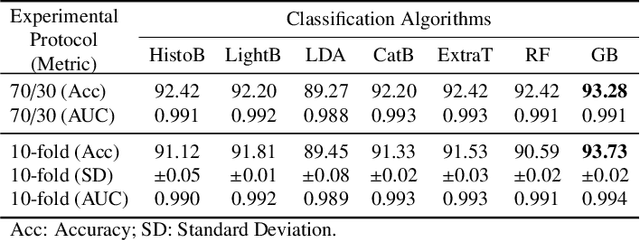
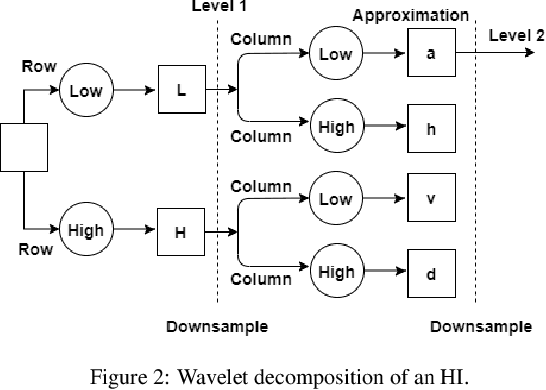
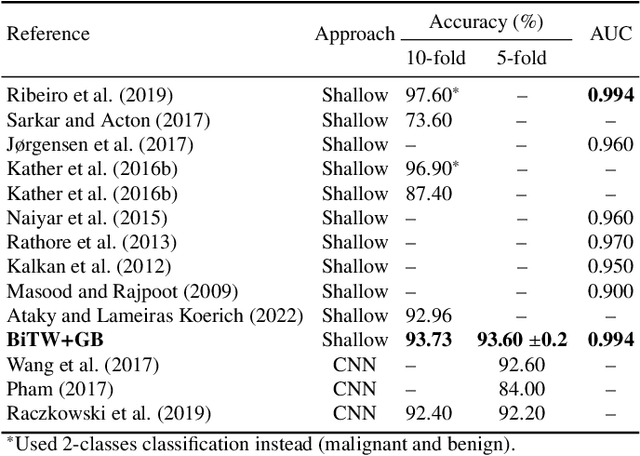
Breast cancer is a health problem that affects mainly the female population. An early detection increases the chances of effective treatment, improving the prognosis of the disease. In this regard, computational tools have been proposed to assist the specialist in interpreting the breast digital image exam, providing features for detecting and diagnosing tumors and cancerous cells. Nonetheless, detecting tumors with a high sensitivity rate and reducing the false positives rate is still challenging. Texture descriptors have been quite popular in medical image analysis, particularly in histopathologic images (HI), due to the variability of both the texture found in such images and the tissue appearance due to irregularity in the staining process. Such variability may exist depending on differences in staining protocol such as fixation, inconsistency in the staining condition, and reagents, either between laboratories or in the same laboratory. Textural feature extraction for quantifying HI information in a discriminant way is challenging given the distribution of intrinsic properties of such images forms a non-deterministic complex system. This paper proposes a method for characterizing texture across HIs with a considerable success rate. By employing ecological diversity measures and discrete wavelet transform, it is possible to quantify the intrinsic properties of such images with promising accuracy on two HI datasets compared with state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge