Jonathan de Matos
Large-Margin Representation Learning for Texture Classification
Jun 17, 2022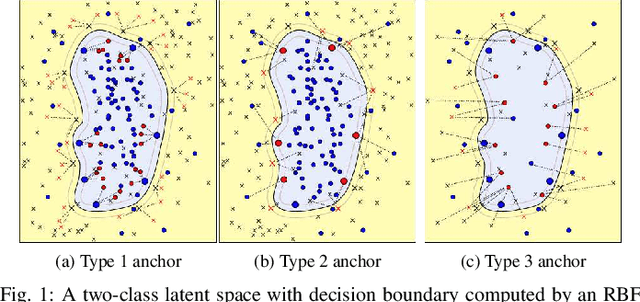
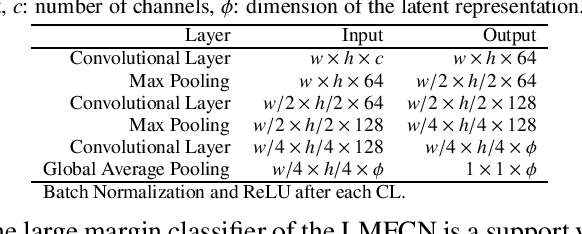
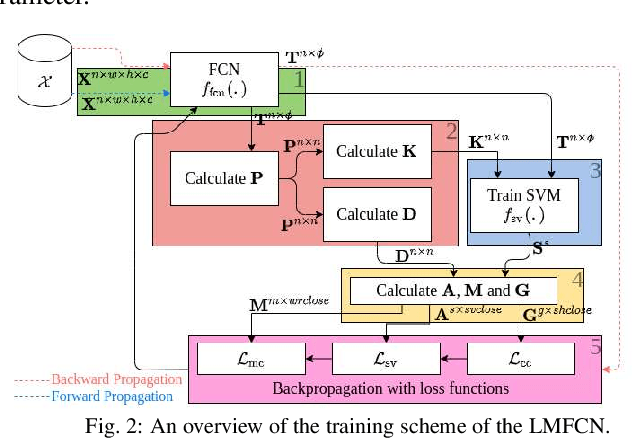
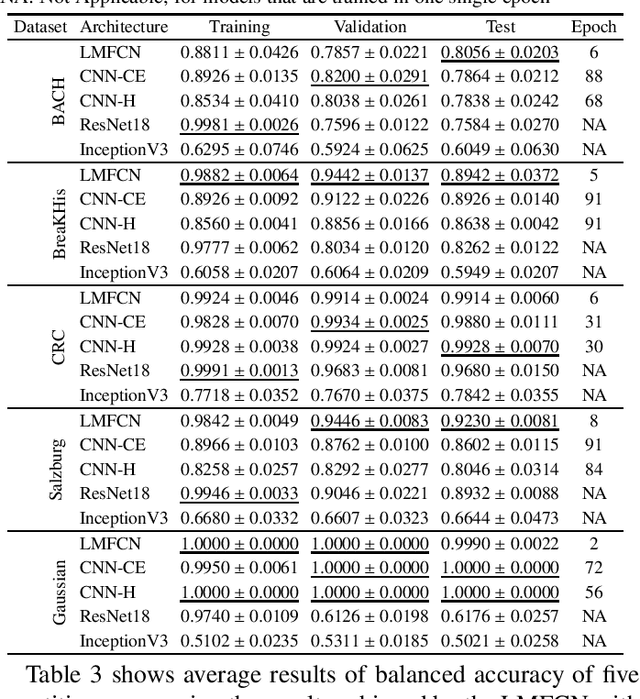
Abstract:This paper presents a novel approach combining convolutional layers (CLs) and large-margin metric learning for training supervised models on small datasets for texture classification. The core of such an approach is a loss function that computes the distances between instances of interest and support vectors. The objective is to update the weights of CLs iteratively to learn a representation with a large margin between classes. Each iteration results in a large-margin discriminant model represented by support vectors based on such a representation. The advantage of the proposed approach w.r.t. convolutional neural networks (CNNs) is two-fold. First, it allows representation learning with a small amount of data due to the reduced number of parameters compared to an equivalent CNN. Second, it has a low training cost since the backpropagation considers only support vectors. The experimental results on texture and histopathologic image datasets have shown that the proposed approach achieves competitive accuracy with lower computational cost and faster convergence when compared to equivalent CNNs.
Multiscale Analysis for Improving Texture Classification
Apr 21, 2022
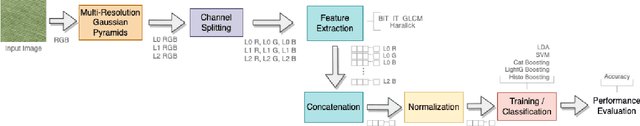
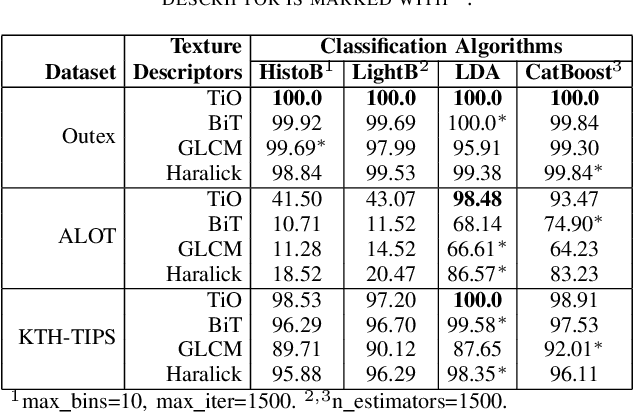
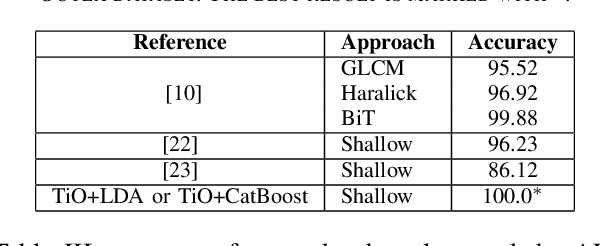
Abstract:Information from an image occurs over multiple and distinct spatial scales. Image pyramid multiresolution representations are a useful data structure for image analysis and manipulation over a spectrum of spatial scales. This paper employs the Gaussian-Laplacian pyramid to treat different spatial frequency bands of a texture separately. First, we generate three images corresponding to three levels of the Gaussian-Laplacian pyramid for an input image to capture intrinsic details. Then we aggregate features extracted from gray and color texture images using bio-inspired texture descriptors, information-theoretic measures, gray-level co-occurrence matrix features, and Haralick statistical features into a single feature vector. Such an aggregation aims at producing features that characterize textures to their maximum extent, unlike employing each descriptor separately, which may lose some relevant textural information and reduce the classification performance. The experimental results on texture and histopathologic image datasets have shown the advantages of the proposed method compared to state-of-the-art approaches. Such findings emphasize the importance of multiscale image analysis and corroborate that the descriptors mentioned above are complementary.
Machine Learning Methods for Histopathological Image Analysis: A Review
Feb 07, 2021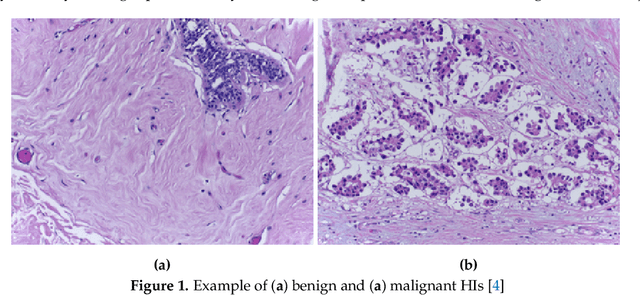
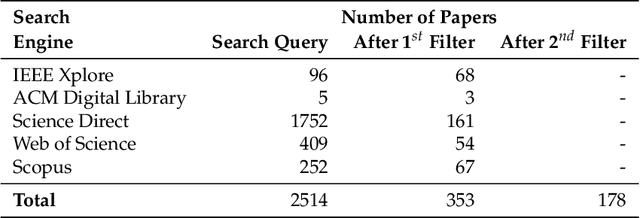
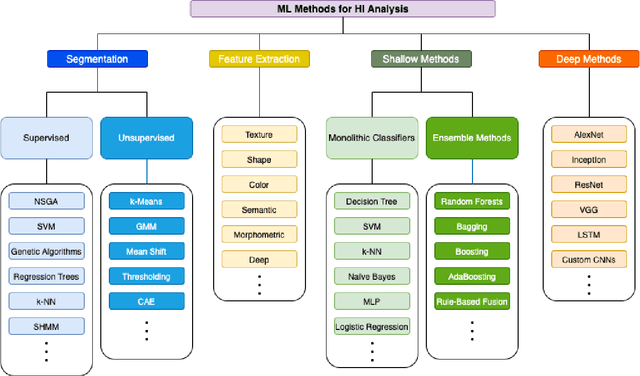
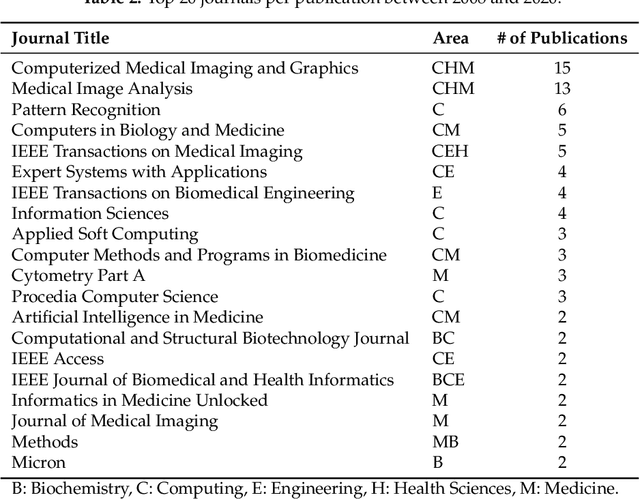
Abstract:Histopathological images (HIs) are the gold standard for evaluating some types of tumors for cancer diagnosis. The analysis of such images is not only time and resource consuming, but also very challenging even for experienced pathologists, resulting in inter- and intra-observer disagreements. One of the ways of accelerating such an analysis is to use computer-aided diagnosis (CAD) systems. In this paper, we present a review on machine learning methods for histopathological image analysis, including shallow and deep learning methods. We also cover the most common tasks in HI analysis, such as segmentation and feature extraction. In addition, we present a list of publicly available and private datasets that have been used in HI research.
Data Augmentation for Histopathological Images Based on Gaussian-Laplacian Pyramid Blending
Jan 31, 2020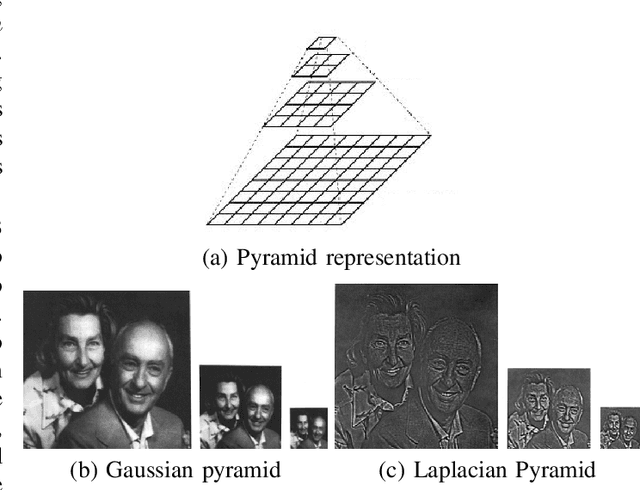
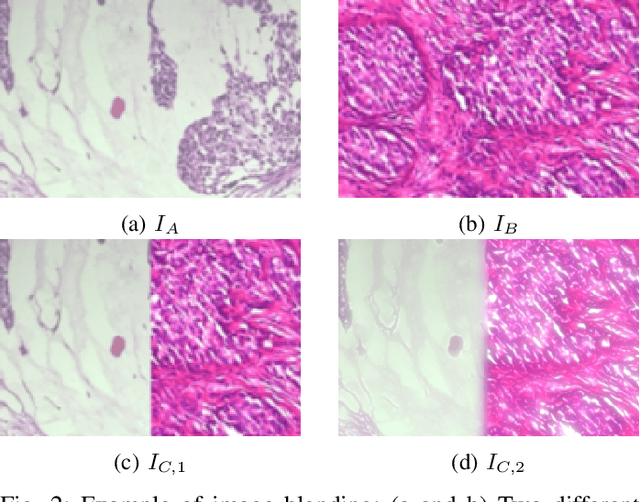
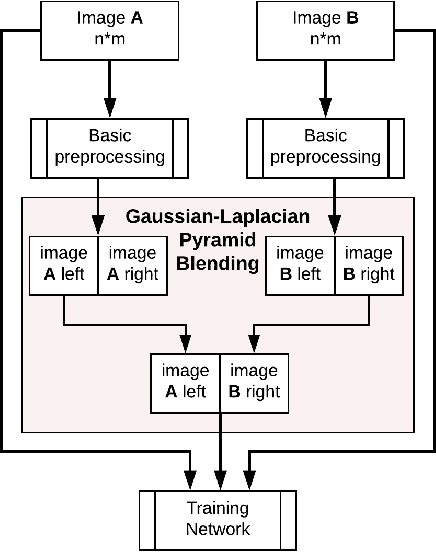
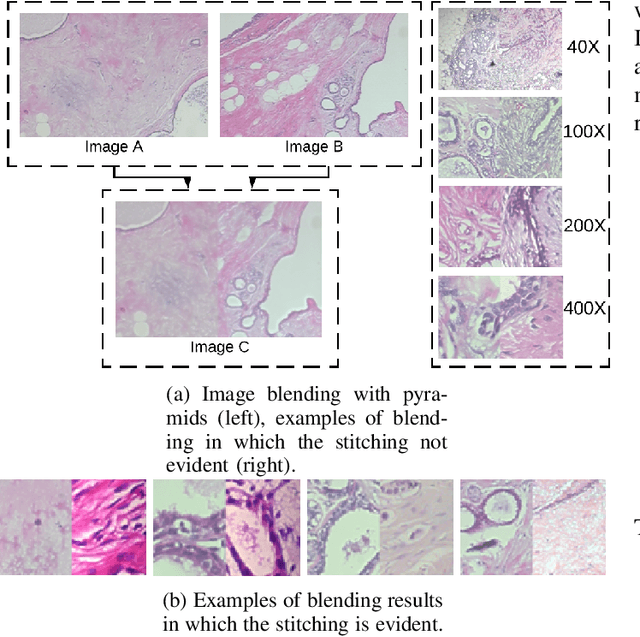
Abstract:Data imbalance is a major problem that affects several machine learning algorithms. Such problems are troublesome because most of the learning algorithms attempts to optimize a loss function based on error measures that do not take into account the data imbalance. Accordingly, the learning algorithm simply generates a trivial model that is biased toward predicting the most frequent class in the training data. Data augmentation techniques have been used to mitigate the data imbalance problem. However, in the case of histopathologic images (HIs), low-level as well as high-level data augmentation techniques still present performance issues when applied in the presence of inter-patient variability; whence the model tends to learn color representations, which are in fact related to the stain process. In this paper, we propose an approach capable of not only augmenting HIs database but also distributing the inter-patient variability by means of image blending using Gaussian-Laplacian pyramid. The proposed approach consists in finding the Gaussian pyramids of two images of different patients and finding the Laplacian pyramids thereof. Afterwards, the left half of one image and the right half of another are joined in each level of Laplacian pyramid, and from the joint pyramids, the original image is reconstructed. This composition, resulting from the blending process, combines stain variation of two patients, avoiding that color misleads the learning process. Experimental results on the BreakHis dataset have shown promising gains vis-\`a-vis the majority of traditional techniques presented in the literature.
Texture CNN for Histopathological Image Classification
May 28, 2019
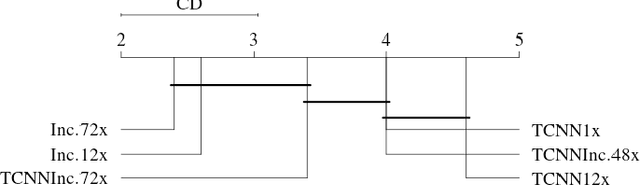
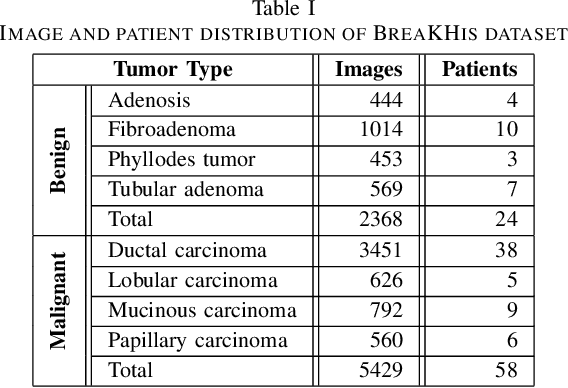
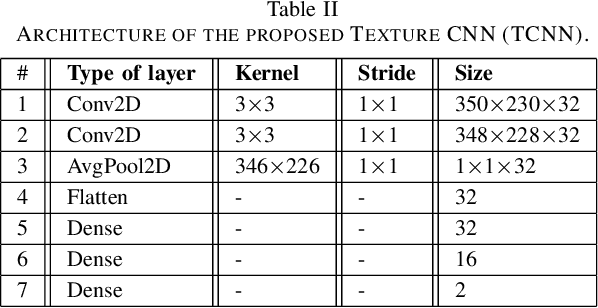
Abstract:Biopsies are the gold standard for breast cancer diagnosis. This task can be improved by the use of Computer Aided Diagnosis (CAD) systems, reducing the time of diagnosis and reducing the inter and intra-observer variability. The advances in computing have brought this type of system closer to reality. However, datasets of Histopathological Images (HI) from biopsies are quite small and unbalanced what makes difficult to use modern machine learning techniques such as deep learning. In this paper we propose a compact architecture based on texture filters that has fewer parameters than traditional deep models but is able to capture the difference between malignant and benign tissues with relative accuracy. The experimental results on the BreakHis dataset have show that the proposed texture CNN achieves almost 90% of accuracy for classifying benign and malignant tissues.
A Novel Orthogonal Direction Mesh Adaptive Direct Search Approach for SVM Hyperparameter Tuning
Apr 26, 2019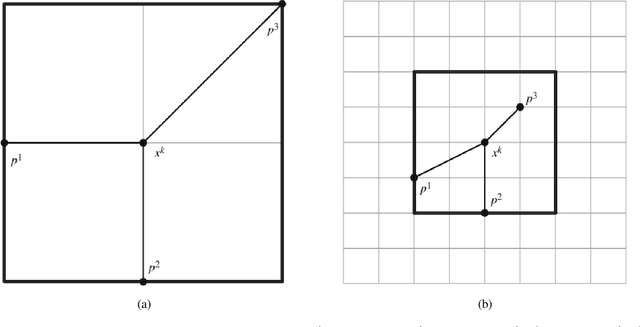
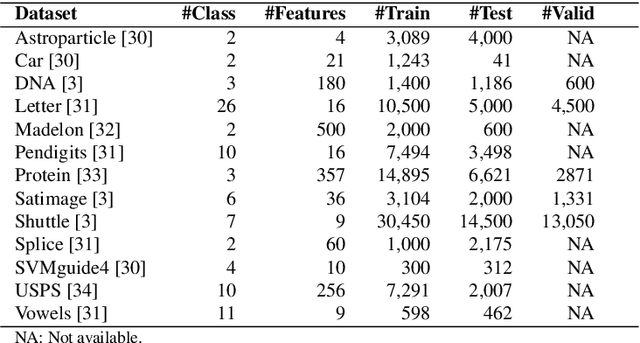
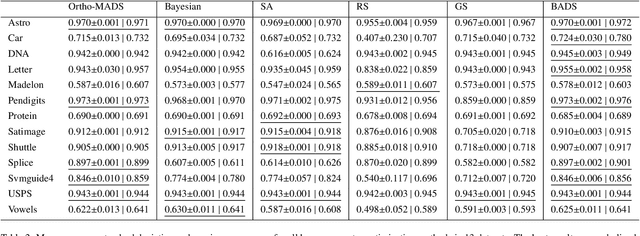
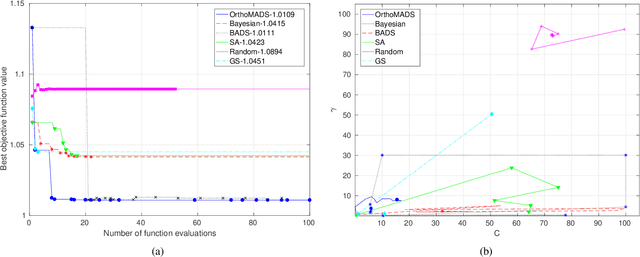
Abstract:In this paper, we propose the use of a black-box optimization method called deterministic Mesh Adaptive Direct Search (MADS) algorithm with orthogonal directions (Ortho-MADS) for the selection of hyperparameters of Support Vector Machines with a Gaussian kernel. Different from most of the methods in the literature that exploit the properties of the data or attempt to minimize the accuracy of a validation dataset over the first quadrant of (C, gamma), the Ortho-MADS provides convergence proof. We present the MADS, followed by the Ortho-MADS, the dynamic stopping criterion defined by the MADS mesh size and two different search strategies (Nelder-Mead and Variable Neighborhood Search) that contribute to a competitive convergence rate as well as a mechanism to escape from undesired local minima. We have investigated the practical selection of hyperparameters for the Support Vector Machine with a Gaussian kernel, i.e., properly choose the hyperparameters gamma (bandwidth) and C (trade-off) on several benchmark datasets. The experimental results have shown that the proposed approach for hyperparameter tuning consistently finds comparable or better solutions, when using a common configuration, than other methods. We have also evaluated the accuracy and the number of function evaluations of the Ortho-MADS with the Nelder-Mead search strategy and the Variable Neighborhood Search strategy using the mesh size as a stopping criterion, and we have achieved accuracy that no other method for hyperparameters optimization could reach.
Histopathologic Image Processing: A Review
Apr 16, 2019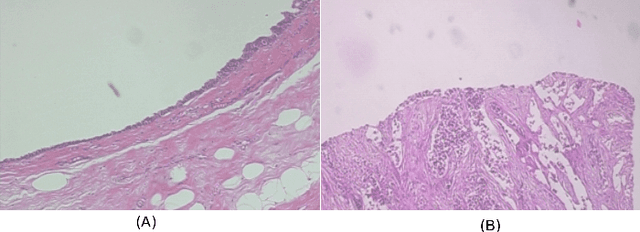
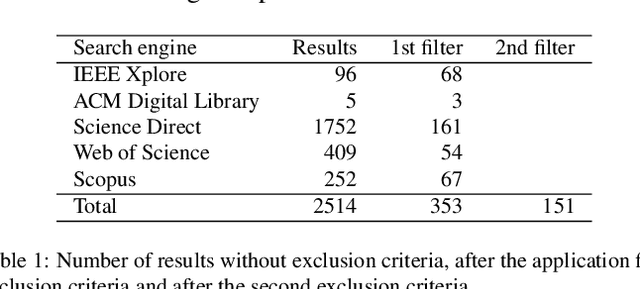

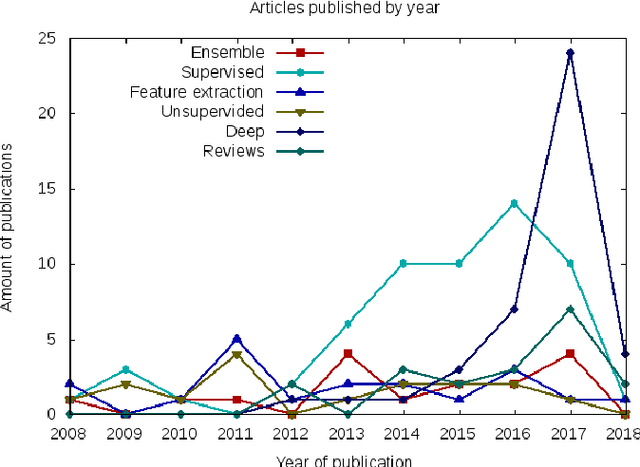
Abstract:Histopathologic Images (HI) are the gold standard for evaluation of some tumors. However, the analysis of such images is challenging even for experienced pathologists, resulting in problems of inter and intra observer. Besides that, the analysis is time and resource consuming. One of the ways to accelerate such an analysis is by using Computer Aided Diagnosis systems. In this work we present a literature review about the computing techniques to process HI, including shallow and deep methods. We cover the most common tasks for processing HI such as segmentation, feature extraction, unsupervised learning and supervised learning. A dataset section show some datasets found during the literature review. We also bring a study case of breast cancer classification using a mix of deep and shallow machine learning methods. The proposed method obtained an accuracy of 91% in the best case, outperforming the compared baseline of the dataset.
Double Transfer Learning for Breast Cancer Histopathologic Image Classification
Apr 16, 2019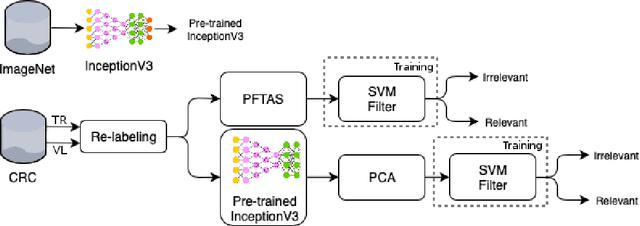
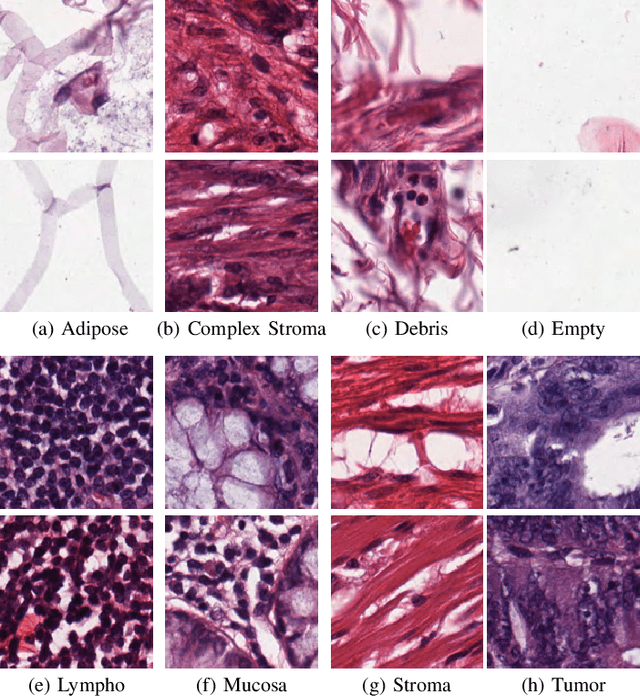
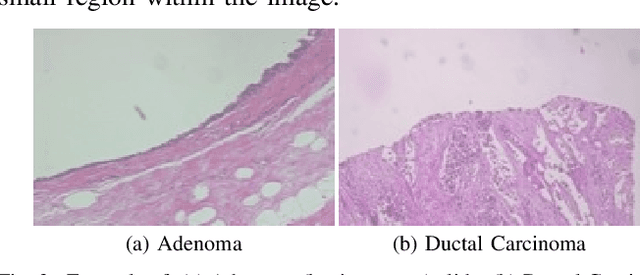
Abstract:This work proposes a classification approach for breast cancer histopathologic images (HI) that uses transfer learning to extract features from HI using an Inception-v3 CNN pre-trained with ImageNet dataset. We also use transfer learning on training a support vector machine (SVM) classifier on a tissue labeled colorectal cancer dataset aiming to filter the patches from a breast cancer HI and remove the irrelevant ones. We show that removing irrelevant patches before training a second SVM classifier, improves the accuracy for classifying malign and benign tumors on breast cancer images. We are able to improve the classification accuracy in 3.7% using the feature extraction transfer learning and an additional 0.7% using the irrelevant patch elimination. The proposed approach outperforms the state-of-the-art in three out of the four magnification factors of the breast cancer dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge