Data Augmentation for Histopathological Images Based on Gaussian-Laplacian Pyramid Blending
Paper and Code
Jan 31, 2020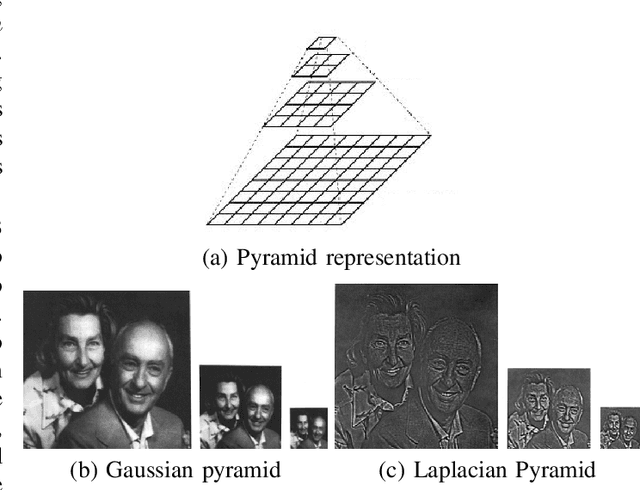
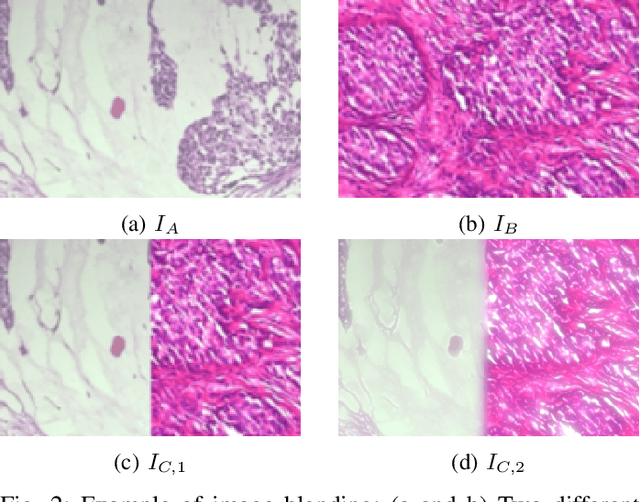
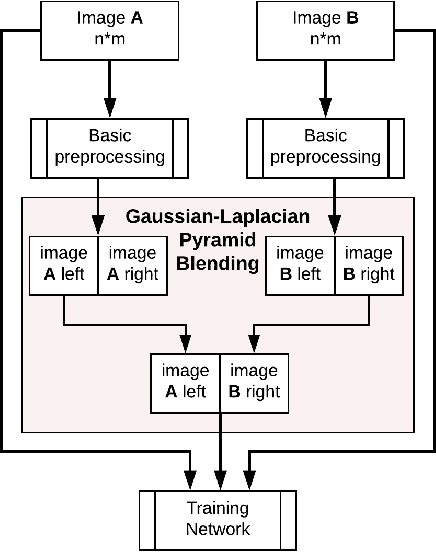
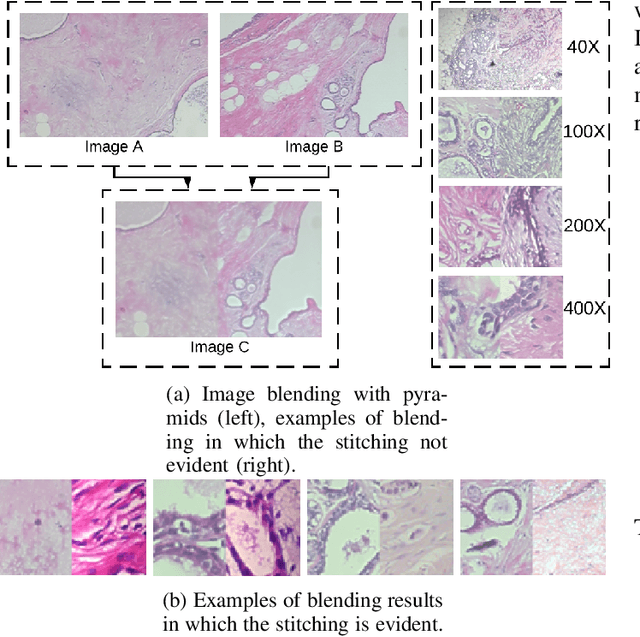
Data imbalance is a major problem that affects several machine learning algorithms. Such problems are troublesome because most of the learning algorithms attempts to optimize a loss function based on error measures that do not take into account the data imbalance. Accordingly, the learning algorithm simply generates a trivial model that is biased toward predicting the most frequent class in the training data. Data augmentation techniques have been used to mitigate the data imbalance problem. However, in the case of histopathologic images (HIs), low-level as well as high-level data augmentation techniques still present performance issues when applied in the presence of inter-patient variability; whence the model tends to learn color representations, which are in fact related to the stain process. In this paper, we propose an approach capable of not only augmenting HIs database but also distributing the inter-patient variability by means of image blending using Gaussian-Laplacian pyramid. The proposed approach consists in finding the Gaussian pyramids of two images of different patients and finding the Laplacian pyramids thereof. Afterwards, the left half of one image and the right half of another are joined in each level of Laplacian pyramid, and from the joint pyramids, the original image is reconstructed. This composition, resulting from the blending process, combines stain variation of two patients, avoiding that color misleads the learning process. Experimental results on the BreakHis dataset have shown promising gains vis-\`a-vis the majority of traditional techniques presented in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge