Lucas Morin
SubGrapher: Visual Fingerprinting of Chemical Structures
Apr 28, 2025Abstract:Automatic extraction of chemical structures from scientific literature plays a crucial role in accelerating research across fields ranging from drug discovery to materials science. Patent documents, in particular, contain molecular information in visual form, which is often inaccessible through traditional text-based searches. In this work, we introduce SubGrapher, a method for the visual fingerprinting of chemical structure images. Unlike conventional Optical Chemical Structure Recognition (OCSR) models that attempt to reconstruct full molecular graphs, SubGrapher focuses on extracting molecular fingerprints directly from chemical structure images. Using learning-based instance segmentation, SubGrapher identifies functional groups and carbon backbones, constructing a substructure-based fingerprint that enables chemical structure retrieval. Our approach is evaluated against state-of-the-art OCSR and fingerprinting methods, demonstrating superior retrieval performance and robustness across diverse molecular depictions. The dataset, models, and code will be made publicly available.
MarkushGrapher: Joint Visual and Textual Recognition of Markush Structures
Mar 20, 2025Abstract:The automated analysis of chemical literature holds promise to accelerate discovery in fields such as material science and drug development. In particular, search capabilities for chemical structures and Markush structures (chemical structure templates) within patent documents are valuable, e.g., for prior-art search. Advancements have been made in the automatic extraction of chemical structures from text and images, yet the Markush structures remain largely unexplored due to their complex multi-modal nature. In this work, we present MarkushGrapher, a multi-modal approach for recognizing Markush structures in documents. Our method jointly encodes text, image, and layout information through a Vision-Text-Layout encoder and an Optical Chemical Structure Recognition vision encoder. These representations are merged and used to auto-regressively generate a sequential graph representation of the Markush structure along with a table defining its variable groups. To overcome the lack of real-world training data, we propose a synthetic data generation pipeline that produces a wide range of realistic Markush structures. Additionally, we present M2S, the first annotated benchmark of real-world Markush structures, to advance research on this challenging task. Extensive experiments demonstrate that our approach outperforms state-of-the-art chemistry-specific and general-purpose vision-language models in most evaluation settings. Code, models, and datasets will be available.
SmolDocling: An ultra-compact vision-language model for end-to-end multi-modal document conversion
Mar 14, 2025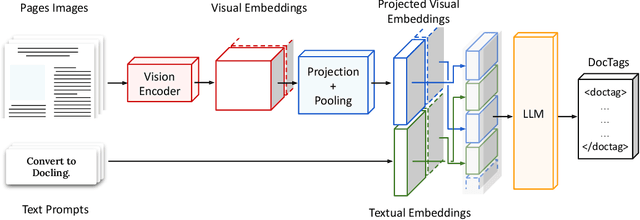

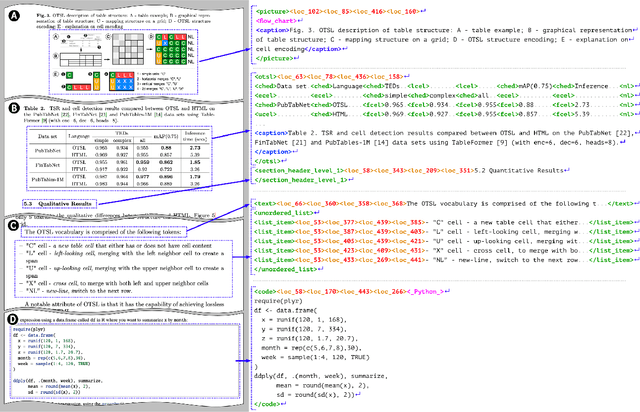
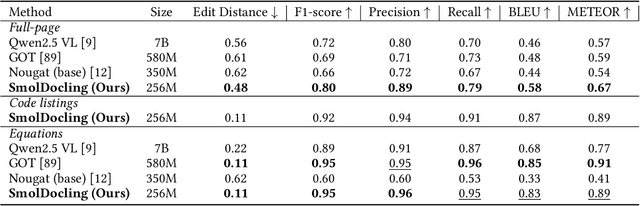
Abstract:We introduce SmolDocling, an ultra-compact vision-language model targeting end-to-end document conversion. Our model comprehensively processes entire pages by generating DocTags, a new universal markup format that captures all page elements in their full context with location. Unlike existing approaches that rely on large foundational models, or ensemble solutions that rely on handcrafted pipelines of multiple specialized models, SmolDocling offers an end-to-end conversion for accurately capturing content, structure and spatial location of document elements in a 256M parameters vision-language model. SmolDocling exhibits robust performance in correctly reproducing document features such as code listings, tables, equations, charts, lists, and more across a diverse range of document types including business documents, academic papers, technical reports, patents, and forms -- significantly extending beyond the commonly observed focus on scientific papers. Additionally, we contribute novel publicly sourced datasets for charts, tables, equations, and code recognition. Experimental results demonstrate that SmolDocling competes with other Vision Language Models that are up to 27 times larger in size, while reducing computational requirements substantially. The model is currently available, datasets will be publicly available soon.
Docling: An Efficient Open-Source Toolkit for AI-driven Document Conversion
Jan 27, 2025



Abstract:We introduce Docling, an easy-to-use, self-contained, MIT-licensed, open-source toolkit for document conversion, that can parse several types of popular document formats into a unified, richly structured representation. It is powered by state-of-the-art specialized AI models for layout analysis (DocLayNet) and table structure recognition (TableFormer), and runs efficiently on commodity hardware in a small resource budget. Docling is released as a Python package and can be used as a Python API or as a CLI tool. Docling's modular architecture and efficient document representation make it easy to implement extensions, new features, models, and customizations. Docling has been already integrated in other popular open-source frameworks (e.g., LangChain, LlamaIndex, spaCy), making it a natural fit for the processing of documents and the development of high-end applications. The open-source community has fully engaged in using, promoting, and developing for Docling, which gathered 10k stars on GitHub in less than a month and was reported as the No. 1 trending repository in GitHub worldwide in November 2024.
Docling Technical Report
Aug 19, 2024



Abstract:This technical report introduces Docling, an easy to use, self-contained, MIT-licensed open-source package for PDF document conversion. It is powered by state-of-the-art specialized AI models for layout analysis (DocLayNet) and table structure recognition (TableFormer), and runs efficiently on commodity hardware in a small resource budget. The code interface allows for easy extensibility and addition of new features and models.
ESG Accountability Made Easy: DocQA at Your Service
Nov 30, 2023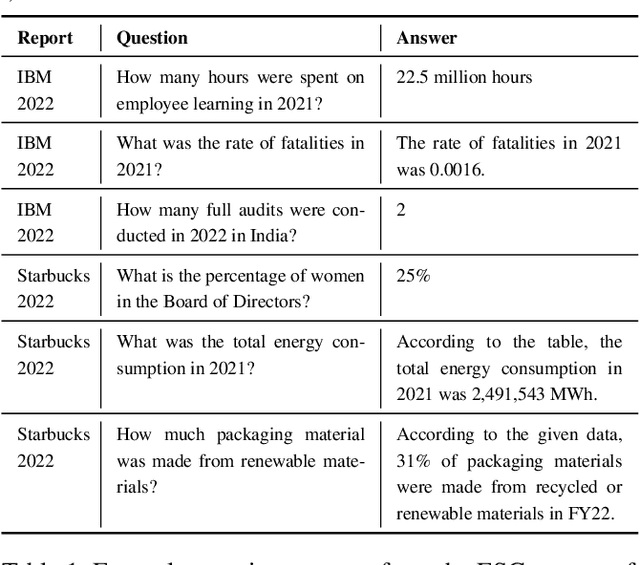
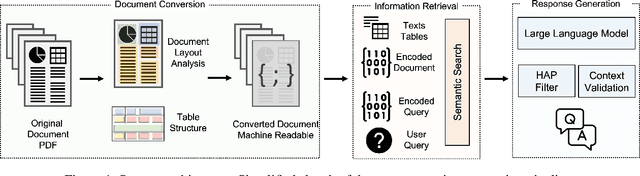
Abstract:We present Deep Search DocQA. This application enables information extraction from documents via a question-answering conversational assistant. The system integrates several technologies from different AI disciplines consisting of document conversion to machine-readable format (via computer vision), finding relevant data (via natural language processing), and formulating an eloquent response (via large language models). Users can explore over 10,000 Environmental, Social, and Governance (ESG) disclosure reports from over 2000 corporations. The Deep Search platform can be accessed at: https://ds4sd.github.io.
MolGrapher: Graph-based Visual Recognition of Chemical Structures
Aug 23, 2023Abstract:The automatic analysis of chemical literature has immense potential to accelerate the discovery of new materials and drugs. Much of the critical information in patent documents and scientific articles is contained in figures, depicting the molecule structures. However, automatically parsing the exact chemical structure is a formidable challenge, due to the amount of detailed information, the diversity of drawing styles, and the need for training data. In this work, we introduce MolGrapher to recognize chemical structures visually. First, a deep keypoint detector detects the atoms. Second, we treat all candidate atoms and bonds as nodes and put them in a graph. This construct allows a natural graph representation of the molecule. Last, we classify atom and bond nodes in the graph with a Graph Neural Network. To address the lack of real training data, we propose a synthetic data generation pipeline producing diverse and realistic results. In addition, we introduce a large-scale benchmark of annotated real molecule images, USPTO-30K, to spur research on this critical topic. Extensive experiments on five datasets show that our approach significantly outperforms classical and learning-based methods in most settings. Code, models, and datasets are available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge