Valery Weber
Docling: An Efficient Open-Source Toolkit for AI-driven Document Conversion
Jan 27, 2025



Abstract:We introduce Docling, an easy-to-use, self-contained, MIT-licensed, open-source toolkit for document conversion, that can parse several types of popular document formats into a unified, richly structured representation. It is powered by state-of-the-art specialized AI models for layout analysis (DocLayNet) and table structure recognition (TableFormer), and runs efficiently on commodity hardware in a small resource budget. Docling is released as a Python package and can be used as a Python API or as a CLI tool. Docling's modular architecture and efficient document representation make it easy to implement extensions, new features, models, and customizations. Docling has been already integrated in other popular open-source frameworks (e.g., LangChain, LlamaIndex, spaCy), making it a natural fit for the processing of documents and the development of high-end applications. The open-source community has fully engaged in using, promoting, and developing for Docling, which gathered 10k stars on GitHub in less than a month and was reported as the No. 1 trending repository in GitHub worldwide in November 2024.
Docling Technical Report
Aug 19, 2024



Abstract:This technical report introduces Docling, an easy to use, self-contained, MIT-licensed open-source package for PDF document conversion. It is powered by state-of-the-art specialized AI models for layout analysis (DocLayNet) and table structure recognition (TableFormer), and runs efficiently on commodity hardware in a small resource budget. The code interface allows for easy extensibility and addition of new features and models.
MolGrapher: Graph-based Visual Recognition of Chemical Structures
Aug 23, 2023Abstract:The automatic analysis of chemical literature has immense potential to accelerate the discovery of new materials and drugs. Much of the critical information in patent documents and scientific articles is contained in figures, depicting the molecule structures. However, automatically parsing the exact chemical structure is a formidable challenge, due to the amount of detailed information, the diversity of drawing styles, and the need for training data. In this work, we introduce MolGrapher to recognize chemical structures visually. First, a deep keypoint detector detects the atoms. Second, we treat all candidate atoms and bonds as nodes and put them in a graph. This construct allows a natural graph representation of the molecule. Last, we classify atom and bond nodes in the graph with a Graph Neural Network. To address the lack of real training data, we propose a synthetic data generation pipeline producing diverse and realistic results. In addition, we introduce a large-scale benchmark of annotated real molecule images, USPTO-30K, to spur research on this critical topic. Extensive experiments on five datasets show that our approach significantly outperforms classical and learning-based methods in most settings. Code, models, and datasets are available.
An Information Extraction and Knowledge Graph Platform for Accelerating Biochemical Discoveries
Jul 19, 2019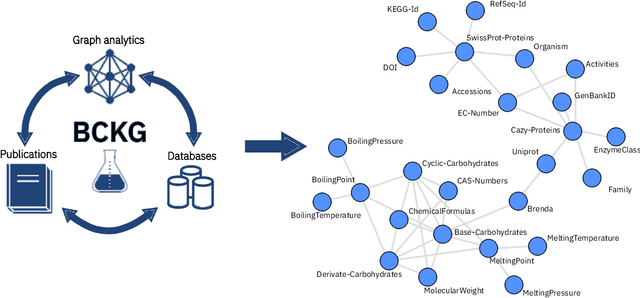
Abstract:Information extraction and data mining in biochemical literature is a daunting task that demands resource-intensive computation and appropriate means to scale knowledge ingestion. Being able to leverage this immense source of technical information helps to drastically reduce costs and time to solution in multiple application fields from food safety to pharmaceutics. We present a scalable document ingestion system that integrates data from databases and publications (in PDF format) in a biochemistry knowledge graph (BCKG). The BCKG is a comprehensive source of knowledge that can be queried to retrieve known biochemical facts and to generate novel insights. After describing the knowledge ingestion framework, we showcase an application of our system in the field of carbohydrate enzymes. The BCKG represents a way to scale knowledge ingestion and automatically exploit prior knowledge to accelerate discovery in biochemical sciences.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge