Laith Alzubaidi
A Review on Deep Learning Autoencoder in the Design of Next-Generation Communication Systems
Dec 18, 2024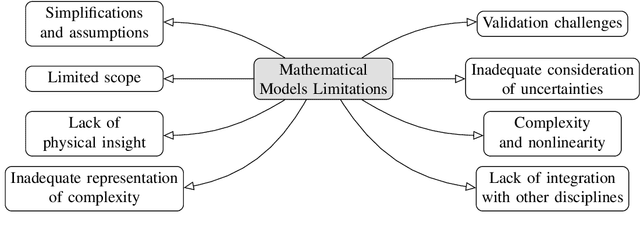
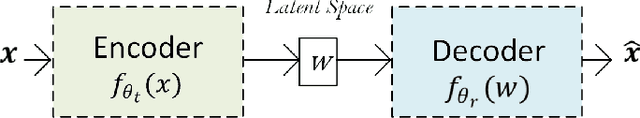
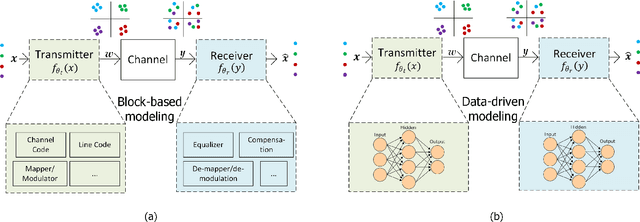
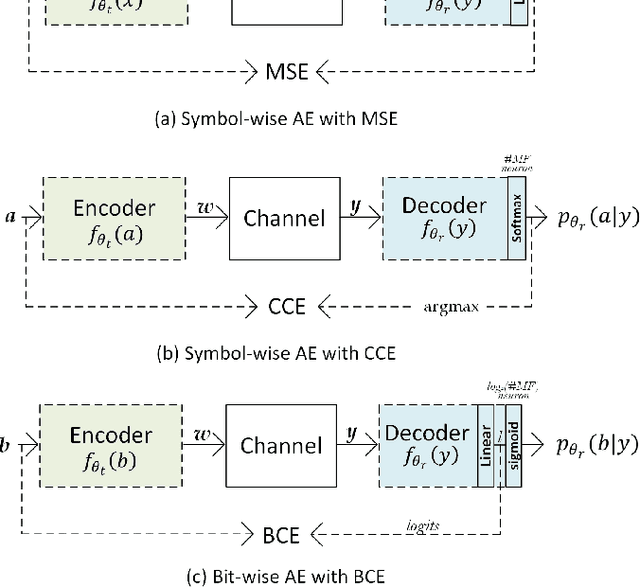
Abstract:Traditional mathematical models used in designing next-generation communication systems often fall short due to inherent simplifications, narrow scope, and computational limitations. In recent years, the incorporation of deep learning (DL) methodologies into communication systems has made significant progress in system design and performance optimisation. Autoencoders (AEs) have become essential, enabling end-to-end learning that allows for the combined optimisation of transmitters and receivers. Consequently, AEs offer a data-driven methodology capable of bridging the gap between theoretical models and real-world complexities. The paper presents a comprehensive survey of the application of AEs within communication systems, with a particular focus on their architectures, associated challenges, and future directions. We examine 120 recent studies across wireless, optical, semantic, and quantum communication fields, categorising them according to transceiver design, channel modelling, digital signal processing, and computational complexity. This paper further examines the challenges encountered in the implementation of AEs, including the need for extensive training data, the risk of overfitting, and the requirement for differentiable channel models. Through data-driven approaches, AEs provide robust solutions for end-to-end system optimisation, surpassing traditional mathematical models confined by simplifying assumptions. This paper also summarises the computational complexity associated with AE-based systems by conducting an in-depth analysis employing the metric of floating-point operations per second (FLOPS). This analysis encompasses the evaluation of matrix multiplications, bias additions, and activation functions. This survey aims to establish a roadmap for future research, emphasising the transformative potential of AEs in the formulation of next-generation communication systems.
Investigating Imperceptibility of Adversarial Attacks on Tabular Data: An Empirical Analysis
Jul 16, 2024Abstract:Adversarial attacks are a potential threat to machine learning models, as they can cause the model to make incorrect predictions by introducing imperceptible perturbations to the input data. While extensively studied in unstructured data like images, their application to structured data like tabular data presents unique challenges due to the heterogeneity and intricate feature interdependencies of tabular data. Imperceptibility in tabular data involves preserving data integrity while potentially causing misclassification, underscoring the need for tailored imperceptibility criteria for tabular data. However, there is currently a lack of standardised metrics for assessing adversarial attacks specifically targeted at tabular data. To address this gap, we derive a set of properties for evaluating the imperceptibility of adversarial attacks on tabular data. These properties are defined to capture seven perspectives of perturbed data: proximity to original inputs, sparsity of alterations, deviation to datapoints in the original dataset, sensitivity of altering sensitive features, immutability of perturbation, feasibility of perturbed values and intricate feature interdepencies among tabular features. Furthermore, we conduct both quantitative empirical evaluation and case-based qualitative examples analysis for seven properties. The evaluation reveals a trade-off between attack success and imperceptibility, particularly concerning proximity, sensitivity, and deviation. Although no evaluated attacks can achieve optimal effectiveness and imperceptibility simultaneously, unbounded attacks prove to be more promised for tabular data in crafting imperceptible adversarial examples. The study also highlights the limitation of evaluated algorithms in controlling sparsity effectively. We suggest incorporating a sparsity metric in future attack design to regulate the number of perturbed features.
Robust and Explainable Framework to Address Data Scarcity in Diagnostic Imaging
Jul 09, 2024Abstract:Deep learning has significantly advanced automatic medical diagnostics and released the occupation of human resources to reduce clinical pressure, yet the persistent challenge of data scarcity in this area hampers its further improvements and applications. To address this gap, we introduce a novel ensemble framework called `Efficient Transfer and Self-supervised Learning based Ensemble Framework' (ETSEF). ETSEF leverages features from multiple pre-trained deep learning models to efficiently learn powerful representations from a limited number of data samples. To the best of our knowledge, ETSEF is the first strategy that combines two pre-training methodologies (Transfer Learning and Self-supervised Learning) with ensemble learning approaches. Various data enhancement techniques, including data augmentation, feature fusion, feature selection, and decision fusion, have also been deployed to maximise the efficiency and robustness of the ETSEF model. Five independent medical imaging tasks, including endoscopy, breast cancer, monkeypox, brain tumour, and glaucoma detection, were tested to demonstrate ETSEF's effectiveness and robustness. Facing limited sample numbers and challenging medical tasks, ETSEF has proved its effectiveness by improving diagnostics accuracies from 10\% to 13.3\% when compared to strong ensemble baseline models and up to 14.4\% improvements compared with published state-of-the-art methods. Moreover, we emphasise the robustness and trustworthiness of the ETSEF method through various vision-explainable artificial intelligence techniques, including Grad-CAM, SHAP, and t-SNE. Compared to those large-scale deep learning models, ETSEF can be deployed flexibly and maintain superior performance for challenging medical imaging tasks, showing the potential to be applied to more areas that lack training data
An Experimental Comparison of Transfer Learning against Self-supervised Learning
Jul 08, 2024Abstract:Recently, transfer learning and self-supervised learning have gained significant attention within the medical field due to their ability to mitigate the challenges posed by limited data availability, improve model generalisation, and reduce computational expenses. Transfer learning and self-supervised learning hold immense potential for advancing medical research. However, it is crucial to recognise that transfer learning and self-supervised learning architectures exhibit distinct advantages and limitations, manifesting variations in accuracy, training speed, and robustness. This paper compares the performance and robustness of transfer learning and self-supervised learning in the medical field. Specifically, we pre-trained two models using the same source domain datasets with different pre-training methods and evaluated them on small-sized medical datasets to identify the factors influencing their final performance. We tested data with several common issues in medical domains, such as data imbalance, data scarcity, and domain mismatch, through comparison experiments to understand their impact on specific pre-trained models. Finally, we provide recommendations to help users apply transfer learning and self-supervised learning methods in medical areas, and build more convenient and efficient deployment strategies.
Physics-informed radial basis network : A local approximating neural network for solving nonlinear PDEs
Apr 20, 2023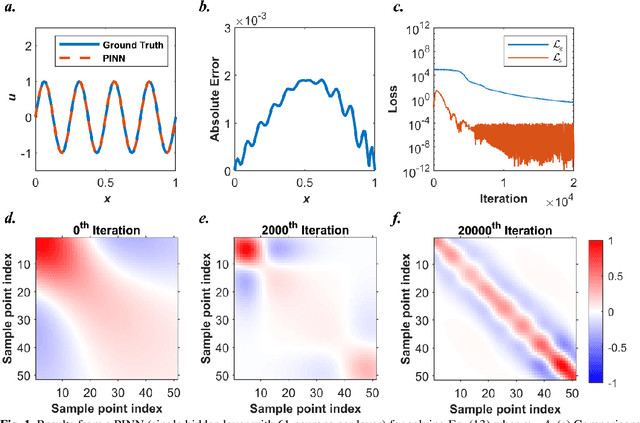
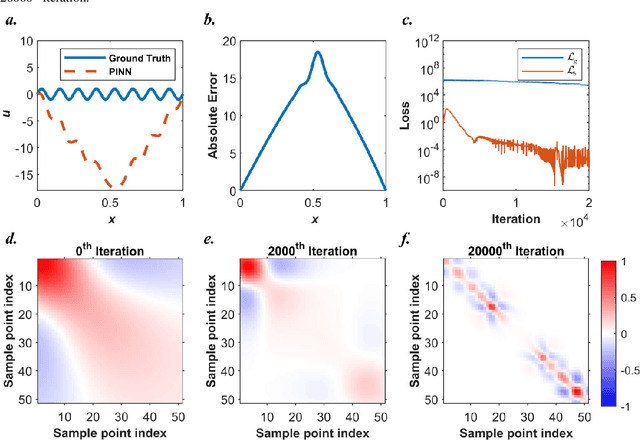
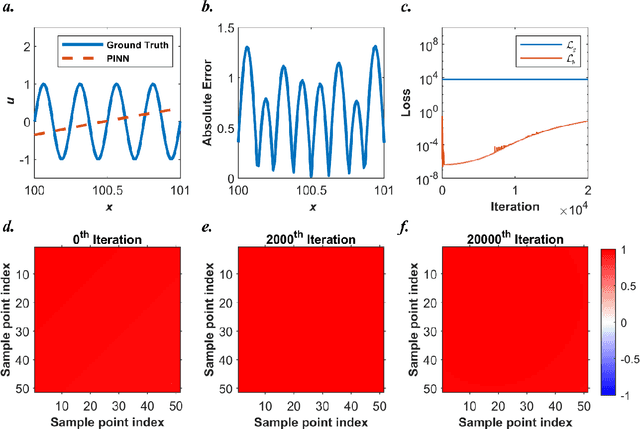
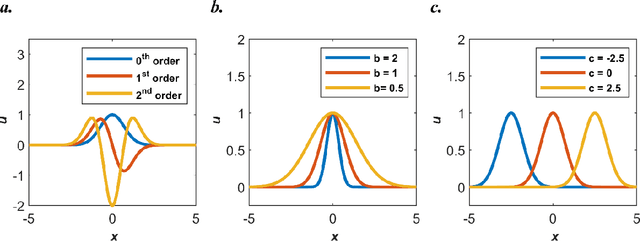
Abstract:Our recent intensive study has found that physics-informed neural networks (PINN) tend to be local approximators after training. This observation leads to this novel physics-informed radial basis network (PIRBN), which can maintain the local property throughout the entire training process. Compared to deep neural networks, a PIRBN comprises of only one hidden layer and a radial basis "activation" function. Under appropriate conditions, we demonstrated that the training of PIRBNs using gradient descendent methods can converge to Gaussian processes. Besides, we studied the training dynamics of PIRBN via the neural tangent kernel (NTK) theory. In addition, comprehensive investigations regarding the initialisation strategies of PIRBN were conducted. Based on numerical examples, PIRBN has been demonstrated to be more effective and efficient than PINN in solving PDEs with high-frequency features and ill-posed computational domains. Moreover, the existing PINN numerical techniques, such as adaptive learning, decomposition and different types of loss functions, are applicable to PIRBN. The programs that can regenerate all numerical results can be found at https://github.com/JinshuaiBai/PIRBN.
Physics-guided deep learning for data scarcity
Nov 24, 2022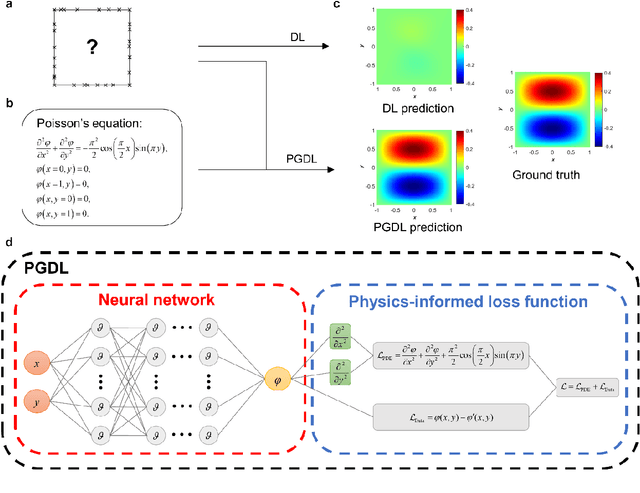
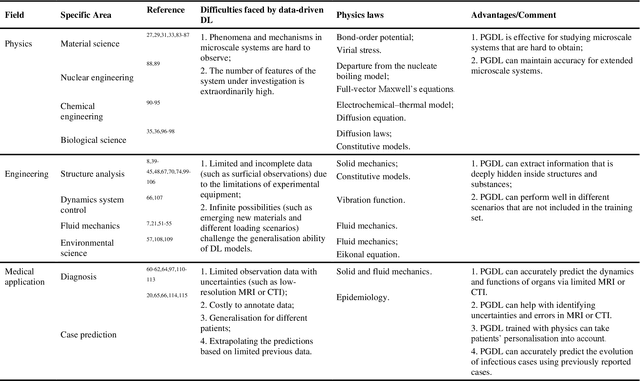
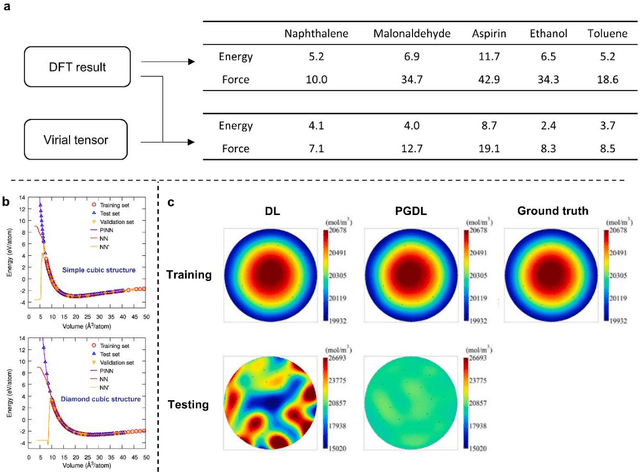
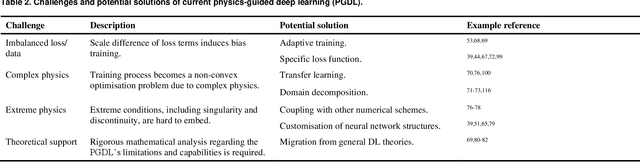
Abstract:Data are the core of deep learning (DL), and the quality of data significantly affects the performance of DL models. However, high-quality and well-annotated databases are hard or even impossible to acquire for use in many applications, such as structural risk estimation and medical diagnosis, which is an essential barrier that blocks the applications of DL in real life. Physics-guided deep learning (PGDL) is a novel type of DL that can integrate physics laws to train neural networks. It can be used for any systems that are controlled or governed by physics laws, such as mechanics, finance and medical applications. It has been shown that, with the additional information provided by physics laws, PGDL achieves great accuracy and generalisation when facing data scarcity. In this review, the details of PGDL are elucidated, and a structured overview of PGDL with respect to data scarcity in various applications is presented, including physics, engineering and medical applications. Moreover, the limitations and opportunities for current PGDL in terms of data scarcity are identified, and the future outlook for PGDL is discussed in depth.
MedNet: Pre-trained Convolutional Neural Network Model for the Medical Imaging Tasks
Oct 13, 2021
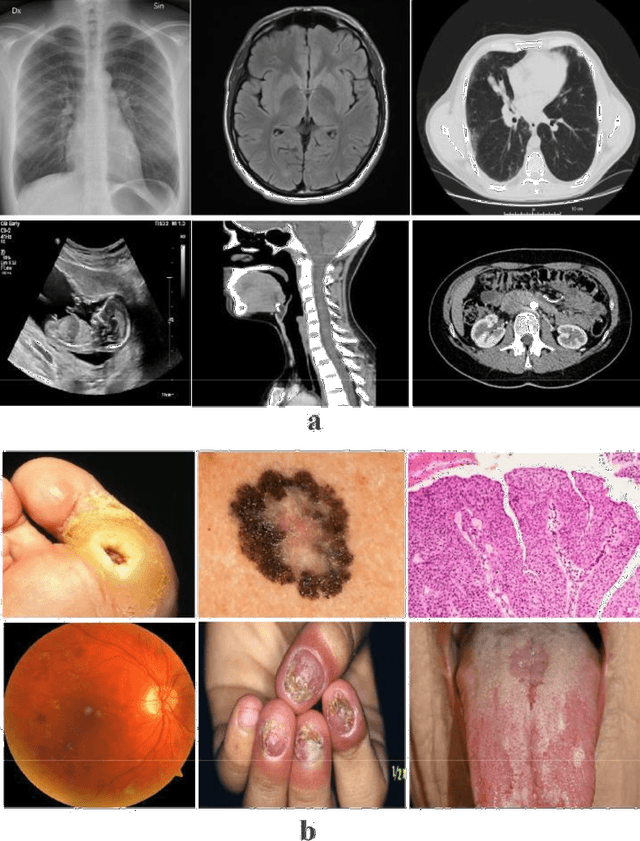
Abstract:Deep Learning (DL) requires a large amount of training data to provide quality outcomes. However, the field of medical imaging suffers from the lack of sufficient data for properly training DL models because medical images require manual labelling carried out by clinical experts thus the process is time-consuming, expensive, and error-prone. Recently, transfer learning (TL) was introduced to reduce the need for the annotation procedure by means of transferring the knowledge performed by a previous task and then fine-tuning the result using a relatively small dataset. Nowadays, multiple classification methods from medical imaging make use of TL from general-purpose pre-trained models, e.g., ImageNet, which has been proven to be ineffective due to the mismatch between the features learned from natural images (ImageNet) and those more specific from medical images especially medical gray images such as X-rays. ImageNet does not have grayscale images such as MRI, CT, and X-ray. In this paper, we propose a novel DL model to be used for addressing classification tasks of medical imaging, called MedNet. To do so, we aim to issue two versions of MedNet. The first one is Gray-MedNet which will be trained on 3M publicly available gray-scale medical images including MRI, CT, X-ray, ultrasound, and PET. The second version is Color-MedNet which will be trained on 3M publicly available color medical images including histopathology, taken images, and many others. To validate the effectiveness MedNet, both versions will be fine-tuned to train on the target tasks of a more reduced set of medical images. MedNet performs as the pre-trained model to tackle any real-world application from medical imaging and achieve the level of generalization needed for dealing with medical imaging tasks, e.g. classification. MedNet would serve the research community as a baseline for future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge