Ali H. Al-Timemy
MedNet: Pre-trained Convolutional Neural Network Model for the Medical Imaging Tasks
Oct 13, 2021
Abstract:Deep Learning (DL) requires a large amount of training data to provide quality outcomes. However, the field of medical imaging suffers from the lack of sufficient data for properly training DL models because medical images require manual labelling carried out by clinical experts thus the process is time-consuming, expensive, and error-prone. Recently, transfer learning (TL) was introduced to reduce the need for the annotation procedure by means of transferring the knowledge performed by a previous task and then fine-tuning the result using a relatively small dataset. Nowadays, multiple classification methods from medical imaging make use of TL from general-purpose pre-trained models, e.g., ImageNet, which has been proven to be ineffective due to the mismatch between the features learned from natural images (ImageNet) and those more specific from medical images especially medical gray images such as X-rays. ImageNet does not have grayscale images such as MRI, CT, and X-ray. In this paper, we propose a novel DL model to be used for addressing classification tasks of medical imaging, called MedNet. To do so, we aim to issue two versions of MedNet. The first one is Gray-MedNet which will be trained on 3M publicly available gray-scale medical images including MRI, CT, X-ray, ultrasound, and PET. The second version is Color-MedNet which will be trained on 3M publicly available color medical images including histopathology, taken images, and many others. To validate the effectiveness MedNet, both versions will be fine-tuned to train on the target tasks of a more reduced set of medical images. MedNet performs as the pre-trained model to tackle any real-world application from medical imaging and achieve the level of generalization needed for dealing with medical imaging tasks, e.g. classification. MedNet would serve the research community as a baseline for future research.
An Efficient Mixture of Deep and Machine Learning Models for COVID-19 and Tuberculosis Detection Using X-Ray Images in Resource Limited Settings
Jul 16, 2020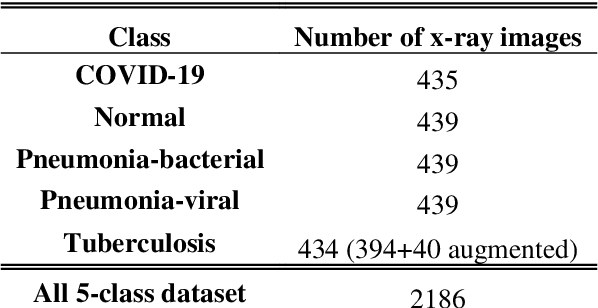
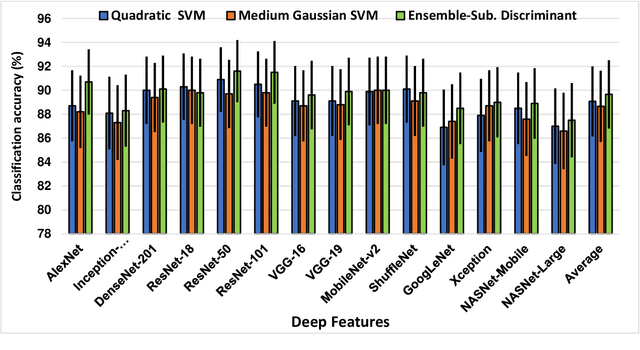
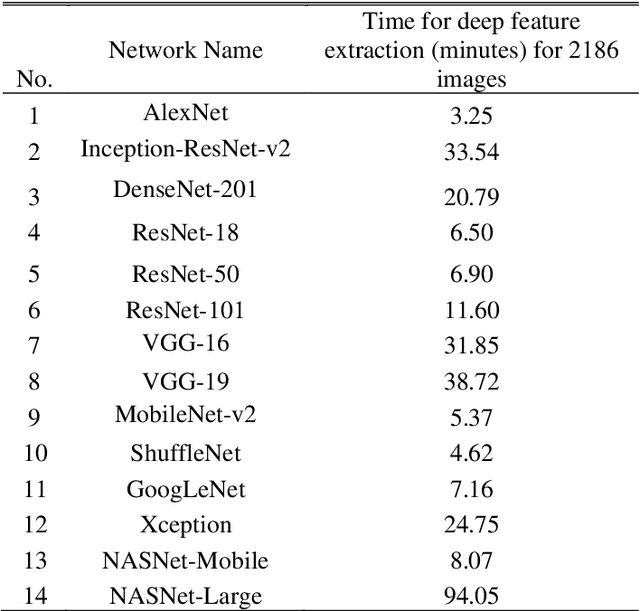
Abstract:Clinicians in the frontline need to assess quickly whether a patient with symptoms indeed has COVID-19 or not. The difficulty of this task is exacerbated in low resource settings that may not have access to biotechnology tests. Furthermore, Tuberculosis (TB) remains a major health problem in several low- and middle-income countries and its common symptoms include fever, cough and tiredness, similarly to COVID-19. In order to help in the detection of COVID-19, we propose the extraction of deep features (DF) from chest X-ray images, a technology available in most hospitals, and their subsequent classification using machine learning methods that do not require large computational resources. We compiled a five-class dataset of X-ray chest images including a balanced number of COVID-19, viral pneumonia, bacterial pneumonia, TB, and healthy cases. We compared the performance of pipelines combining 14 individual state-of-the-art pre-trained deep networks for DF extraction with traditional machine learning classifiers. A pipeline consisting of ResNet-50 for DF computation and ensemble of subspace discriminant classifier was the best performer in the classification of the five classes, achieving a detection accuracy of 91.6+ 2.6% (accuracy + 95% Confidence Interval). Furthermore, the same pipeline achieved accuracies of 98.6+1.4% and 99.9+0.5% in simpler three-class and two-class classification problems focused on distinguishing COVID-19, TB and healthy cases; and COVID-19 and healthy images, respectively. The pipeline was computationally efficient requiring just 0.19 second to extract DF per X-ray image and 2 minutes for training a traditional classifier with more than 2000 images on a CPU machine. The results suggest the potential benefits of using our pipeline in the detection of COVID-19, particularly in resource-limited settings and it can run with limited computational resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge