Javier Escudero
PySeizure: A single machine learning classifier framework to detect seizures in diverse datasets
Aug 10, 2025Abstract:Reliable seizure detection is critical for diagnosing and managing epilepsy, yet clinical workflows remain dependent on time-consuming manual EEG interpretation. While machine learning has shown promise, existing approaches often rely on dataset-specific optimisations, limiting their real-world applicability and reproducibility. Here, we introduce an innovative, open-source machine-learning framework that enables robust and generalisable seizure detection across varied clinical datasets. We evaluate our approach on two publicly available EEG datasets that differ in patient populations and electrode configurations. To enhance robustness, the framework incorporates an automated pre-processing pipeline to standardise data and a majority voting mechanism, in which multiple models independently assess each second of EEG before reaching a final decision. We train, tune, and evaluate models within each dataset, assessing their cross-dataset transferability. Our models achieve high within-dataset performance (AUC 0.904+/-0.059 for CHB-MIT and 0.864+/-0.060 for TUSZ) and demonstrate strong generalisation across datasets despite differences in EEG setups and populations (AUC 0.615+/-0.039 for models trained on CHB-MIT and tested on TUSZ and 0.762+/-0.175 in the reverse case) without any post-processing. Furthermore, a mild post-processing improved the within-dataset results to 0.913+/-0.064 and 0.867+/-0.058 and cross-dataset results to 0.619+/-0.036 and 0.768+/-0.172. These results underscore the potential of, and essential considerations for, deploying our framework in diverse clinical settings. By making our methodology fully reproducible, we provide a foundation for advancing clinically viable, dataset-agnostic seizure detection systems. This approach has the potential for widespread adoption, complementing rather than replacing expert interpretation, and accelerating clinical integration.
Preictal Period Optimization for Deep Learning-Based Epileptic Seizure Prediction
Jul 20, 2024Abstract:Accurate prediction of epileptic seizures could prove critical for improving patient safety and quality of life in drug-resistant epilepsy. Although deep learning-based approaches have shown promising seizure prediction performance using scalp electroencephalogram (EEG) signals, substantial limitations still impede their clinical adoption. Furthermore, identifying the optimal preictal period (OPP) for labeling EEG segments remains a challenge. Here, we not only develop a competitive deep learning model for seizure prediction but, more importantly, leverage it to demonstrate a methodology to comprehensively evaluate the predictive performance in the seizure prediction task. For this, we introduce a CNN-Transformer deep learning model to detect preictal spatiotemporal dynamics, alongside a novel Continuous Input-Output Performance Ratio (CIOPR) metric to determine the OPP. We trained and evaluated our model on 19 pediatric patients of the open-access CHB-MIT dataset in a subject-specific manner. Using the OPP of each patient, preictal and interictal segments were correctly identified with an average sensitivity of 99.31%, specificity of 95.34%, AUC of 99.35%, and F1- score of 97.46%, while prediction time averaged 76.8 minutes before onset. Notably, our novel CIOPR metric allowed outlining the impact of different preictal period definitions on prediction time, accuracy, output stability, and transition time between interictal and preictal states in a comprehensive and quantitative way and highlighted the importance of considering both inter- and intra-patient variability in seizure prediction.
Collaborative learning of common latent representations in routinely collected multivariate ICU physiological signals
Feb 27, 2024Abstract:In Intensive Care Units (ICU), the abundance of multivariate time series presents an opportunity for machine learning (ML) to enhance patient phenotyping. In contrast to previous research focused on electronic health records (EHR), here we propose an ML approach for phenotyping using routinely collected physiological time series data. Our new algorithm integrates Long Short-Term Memory (LSTM) networks with collaborative filtering concepts to identify common physiological states across patients. Tested on real-world ICU clinical data for intracranial hypertension (IH) detection in patients with brain injury, our method achieved an area under the curve (AUC) of 0.889 and average precision (AP) of 0.725. Moreover, our algorithm outperforms autoencoders in learning more structured latent representations of the physiological signals. These findings highlight the promise of our methodology for patient phenotyping, leveraging routinely collected multivariate time series to improve clinical care practices.
Core consistency diagnosis for Block Term Decomposition in rank $(L_r, L_r, 1)$
Dec 18, 2023



Abstract:Determining the underlying number of components $R$ in tensor decompositions is challenging. Diverse techniques exist for various decompositions, notably the core consistency diagnostic (CORCONDIA) for Canonical Polyadic Decomposition (CPD). Here, we propose a model that intuitively adapts CORCONDIA for rank estimation in Block Term Decomposition (BTD) of rank $(L_r, L_r, 1)$: BTDCORCONDIA. Our metric was tested on simulated and real-world tensor data, including assessments of its sensitivity to noise and the indeterminacy of BTD $(L_r, L_r, 1)$. We found that selecting appropriate $R$ and $L_r$ led to core consistency reaching or close to 100%, and BTDCORCONDIA is efficient when the tensor has significantly more elements than the core. Our results confirm that CORCONDIA can be extended to BTD $(L_r, L_r, 1)$, and the resulting metric can assist in the process of determining the number of components in this tensor factorisation.
VAE-IF: Deep feature extraction with averaging for unsupervised artifact detection in routine acquired ICU time-series
Dec 10, 2023Abstract:Artifacts are a common problem in physiological time-series data collected from intensive care units (ICU) and other settings. They affect the quality and reliability of clinical research and patient care. Manual annotation of artifacts is costly and time-consuming, rendering it impractical. Automated methods are desired. Here, we propose a novel unsupervised approach to detect artifacts in clinical-standard minute-by-minute resolution ICU data without any prior labeling or signal-specific knowledge. Our approach combines a variational autoencoder (VAE) and an isolation forest (iForest) model to learn features and identify anomalies in different types of vital signs, such as blood pressure, heart rate, and intracranial pressure. We evaluate our approach on a real-world ICU dataset and compare it with supervised models based on long short-term memory (LSTM) and XGBoost. We show that our approach achieves comparable sensitivity and generalizes well to an external dataset. We also visualize the latent space learned by the VAE and demonstrate its ability to disentangle clean and noisy samples. Our approach offers a promising solution for cleaning ICU data in clinical research and practice without the need for any labels whatsoever.
Hybrid Artifact Detection System for Minute Resolution Blood Pressure Signals from ICU
Mar 11, 2022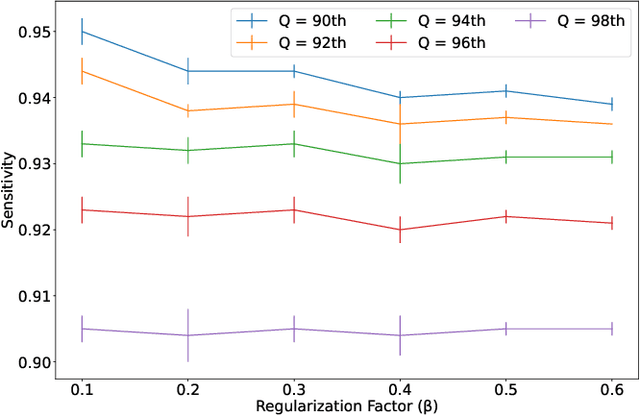

Abstract:Physiological monitoring in intensive care units generates data that can be used to aid clinical decision making facilitating early interventions. However, the low data quality of physiological signals due to the recording conditions in clinical settings limits the automated extraction of relevant information and leads to significant numbers of false alarms. This paper investigates the utilization of a hybrid artifact detection system that combines a Variational Autoencoder with a statistical detection component for the labeling of artifactual samples to automate the costly process of cleaning physiological recordings. The system is applied to mean blood pressure signals from an intensive care unit dataset recorded within the scope of the KidsBrainIT project. Its performance is benchmarked to manual annotations made by trained researchers. Our preliminary results indicate that the system is capable of consistently achieving sensitivity and specificity levels that surpass 90%. Thus, it provides an initial foundation that can be expanded upon to partially automate data cleaning in offline applications and reduce false alarms in online applications.
An Efficient Mixture of Deep and Machine Learning Models for COVID-19 and Tuberculosis Detection Using X-Ray Images in Resource Limited Settings
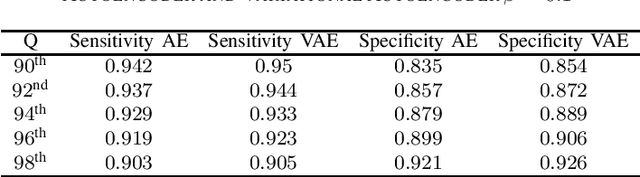
Jul 16, 2020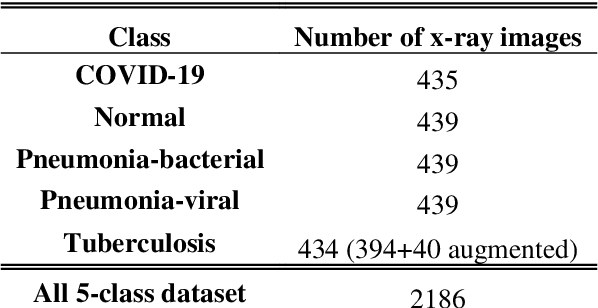
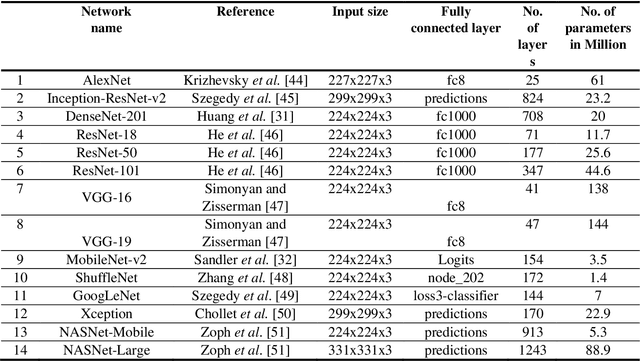
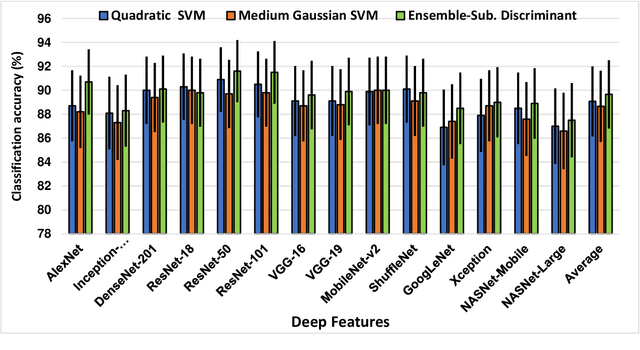
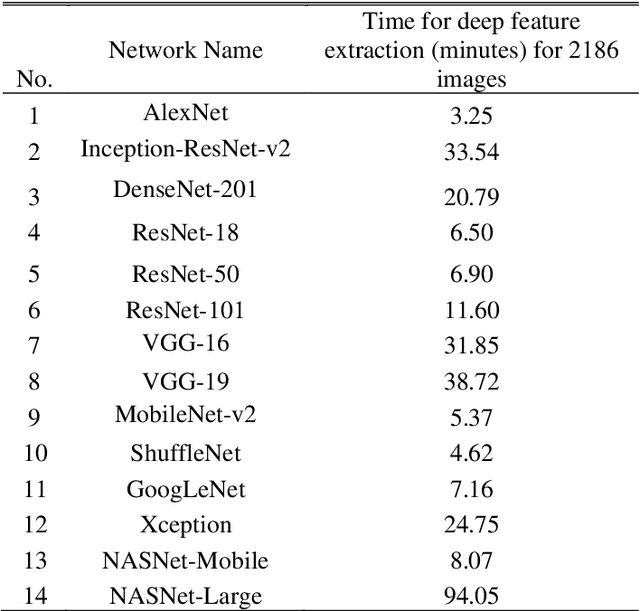
Abstract:Clinicians in the frontline need to assess quickly whether a patient with symptoms indeed has COVID-19 or not. The difficulty of this task is exacerbated in low resource settings that may not have access to biotechnology tests. Furthermore, Tuberculosis (TB) remains a major health problem in several low- and middle-income countries and its common symptoms include fever, cough and tiredness, similarly to COVID-19. In order to help in the detection of COVID-19, we propose the extraction of deep features (DF) from chest X-ray images, a technology available in most hospitals, and their subsequent classification using machine learning methods that do not require large computational resources. We compiled a five-class dataset of X-ray chest images including a balanced number of COVID-19, viral pneumonia, bacterial pneumonia, TB, and healthy cases. We compared the performance of pipelines combining 14 individual state-of-the-art pre-trained deep networks for DF extraction with traditional machine learning classifiers. A pipeline consisting of ResNet-50 for DF computation and ensemble of subspace discriminant classifier was the best performer in the classification of the five classes, achieving a detection accuracy of 91.6+ 2.6% (accuracy + 95% Confidence Interval). Furthermore, the same pipeline achieved accuracies of 98.6+1.4% and 99.9+0.5% in simpler three-class and two-class classification problems focused on distinguishing COVID-19, TB and healthy cases; and COVID-19 and healthy images, respectively. The pipeline was computationally efficient requiring just 0.19 second to extract DF per X-ray image and 2 minutes for training a traditional classifier with more than 2000 images on a CPU machine. The results suggest the potential benefits of using our pipeline in the detection of COVID-19, particularly in resource-limited settings and it can run with limited computational resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge