Koen Dercksen
What Can Natural Language Processing Do for Peer Review?
May 10, 2024



Abstract:The number of scientific articles produced every year is growing rapidly. Providing quality control over them is crucial for scientists and, ultimately, for the public good. In modern science, this process is largely delegated to peer review -- a distributed procedure in which each submission is evaluated by several independent experts in the field. Peer review is widely used, yet it is hard, time-consuming, and prone to error. Since the artifacts involved in peer review -- manuscripts, reviews, discussions -- are largely text-based, Natural Language Processing has great potential to improve reviewing. As the emergence of large language models (LLMs) has enabled NLP assistance for many new tasks, the discussion on machine-assisted peer review is picking up the pace. Yet, where exactly is help needed, where can NLP help, and where should it stand aside? The goal of our paper is to provide a foundation for the future efforts in NLP for peer-reviewing assistance. We discuss peer review as a general process, exemplified by reviewing at AI conferences. We detail each step of the process from manuscript submission to camera-ready revision, and discuss the associated challenges and opportunities for NLP assistance, illustrated by existing work. We then turn to the big challenges in NLP for peer review as a whole, including data acquisition and licensing, operationalization and experimentation, and ethical issues. To help consolidate community efforts, we create a companion repository that aggregates key datasets pertaining to peer review. Finally, we issue a detailed call for action for the scientific community, NLP and AI researchers, policymakers, and funding bodies to help bring the research in NLP for peer review forward. We hope that our work will help set the agenda for research in machine-assisted scientific quality control in the age of AI, within the NLP community and beyond.
REL: An Entity Linker Standing on the Shoulders of Giants
Jun 02, 2020



Abstract:Entity linking is a standard component in modern retrieval system that is often performed by third-party toolkits. Despite the plethora of open source options, it is difficult to find a single system that has a modular architecture where certain components may be replaced, does not depend on external sources, can easily be updated to newer Wikipedia versions, and, most important of all, has state-of-the-art performance. The REL system presented in this paper aims to fill that gap. Building on state-of-the-art neural components from natural language processing research, it is provided as a Python package as well as a web API. We also report on an experimental comparison against both well-established systems and the current state-of-the-art on standard entity linking benchmarks.
Dealing with Label Scarcity in Computational Pathology: A Use Case in Prostate Cancer Classification
May 16, 2019
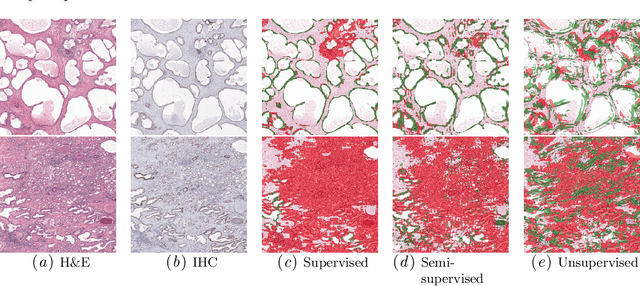
Abstract:Large amounts of unlabelled data are commonplace for many applications in computational pathology, whereas labelled data is often expensive, both in time and cost, to acquire. We investigate the performance of unsupervised and supervised deep learning methods when few labelled data are available. Three methods are compared: clustering autoencoder latent vectors (unsupervised), a single layer classifier combined with a pre-trained autoencoder (semi-supervised), and a supervised CNN. We apply these methods on hematoxylin and eosin (H&E) stained prostatectomy images to classify tumour versus non-tumour tissue. Results show that semi-/unsupervised methods have an advantage over supervised learning when few labels are available. Additionally, we show that incorporating immunohistochemistry (IHC) stained data provides an increase in performance over only using H&E.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge