Junier Oliva
Department of Computer Science, University of North Carolina at Chapel Hill
Towards Universal Neural Inference
Aug 12, 2025Abstract:Real-world data often appears in diverse, disjoint forms -- with varying schemas, inconsistent semantics, and no fixed feature ordering -- making it challenging to build general-purpose models that can leverage information across datasets. We introduce ASPIRE, Arbitrary Set-based Permutation-Invariant Reasoning Engine, a Universal Neural Inference model for semantic reasoning and prediction over heterogeneous structured data. ASPIRE combines a permutation-invariant, set-based Transformer with a semantic grounding module that incorporates natural language descriptions, dataset metadata, and in-context examples to learn cross-dataset feature dependencies. This architecture allows ASPIRE to ingest arbitrary sets of feature--value pairs and support examples, align semantics across disjoint tables, and make predictions for any specified target. Once trained, ASPIRE generalizes to new inference tasks without additional tuning. In addition to delivering strong results across diverse benchmarks, ASPIRE naturally supports cost-aware active feature acquisition in an open-world setting, selecting informative features under test-time budget constraints for an arbitrary unseen dataset. These capabilities position ASPIRE as a step toward truly universal, semantics-aware inference over structured data.
NOCTA: Non-Greedy Objective Cost-Tradeoff Acquisition for Longitudinal Data
Jul 16, 2025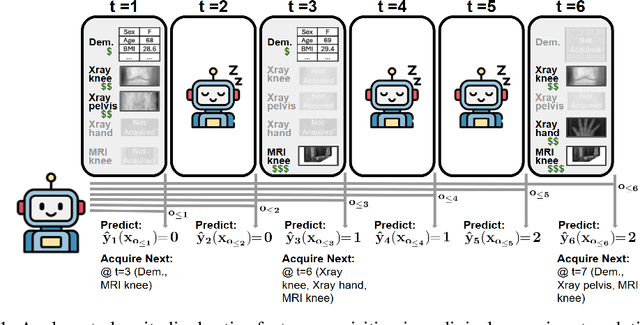
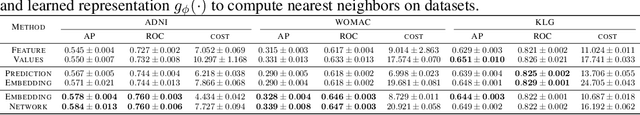
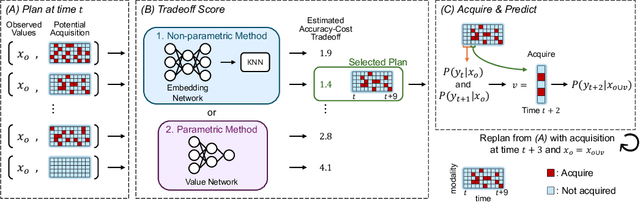
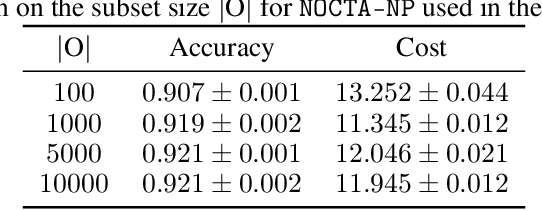
Abstract:In many critical applications, resource constraints limit the amount of information that can be gathered to make predictions. For example, in healthcare, patient data often spans diverse features ranging from lab tests to imaging studies. Each feature may carry different information and must be acquired at a respective cost of time, money, or risk to the patient. Moreover, temporal prediction tasks, where both instance features and labels evolve over time, introduce additional complexity in deciding when or what information is important. In this work, we propose NOCTA, a Non-Greedy Objective Cost-Tradeoff Acquisition method that sequentially acquires the most informative features at inference time while accounting for both temporal dynamics and acquisition cost. We first introduce a cohesive estimation target for our NOCTA setting, and then develop two complementary estimators: 1) a non-parametric method based on nearest neighbors to guide the acquisition (NOCTA-NP), and 2) a parametric method that directly predicts the utility of potential acquisitions (NOCTA-P). Experiments on synthetic and real-world medical datasets demonstrate that both NOCTA variants outperform existing baselines.
VIN-NBV: A View Introspection Network for Next-Best-View Selection for Resource-Efficient 3D Reconstruction
May 09, 2025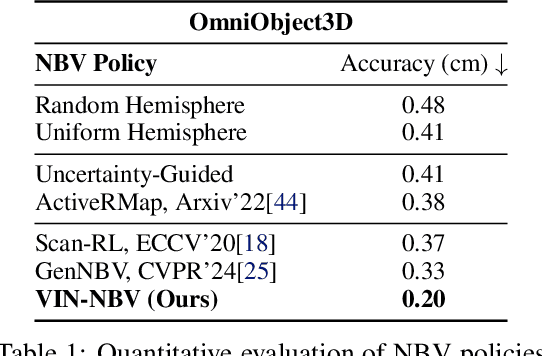
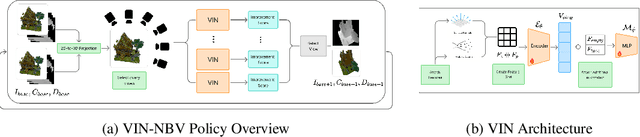
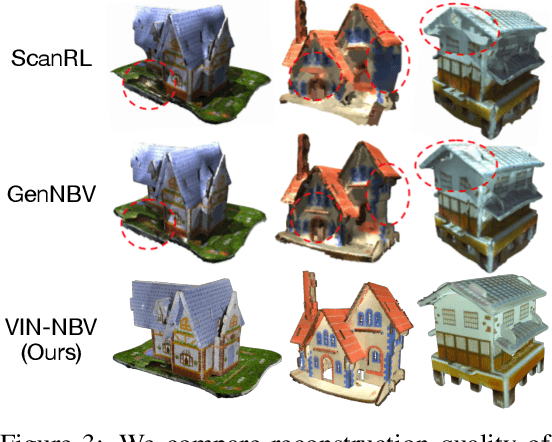
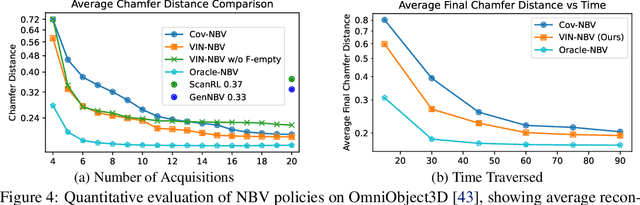
Abstract:Next Best View (NBV) algorithms aim to acquire an optimal set of images using minimal resources, time, or number of captures to enable efficient 3D reconstruction of a scene. Existing approaches often rely on prior scene knowledge or additional image captures and often develop policies that maximize coverage. Yet, for many real scenes with complex geometry and self-occlusions, coverage maximization does not lead to better reconstruction quality directly. In this paper, we propose the View Introspection Network (VIN), which is trained to predict the reconstruction quality improvement of views directly, and the VIN-NBV policy. A greedy sequential sampling-based policy, where at each acquisition step, we sample multiple query views and choose the one with the highest VIN predicted improvement score. We design the VIN to perform 3D-aware featurization of the reconstruction built from prior acquisitions, and for each query view create a feature that can be decoded into an improvement score. We then train the VIN using imitation learning to predict the reconstruction improvement score. We show that VIN-NBV improves reconstruction quality by ~30% over a coverage maximization baseline when operating with constraints on the number of acquisitions or the time in motion.
Distribution Guided Active Feature Acquisition
Oct 04, 2024



Abstract:Human agents routinely reason on instances with incomplete and muddied data (and weigh the cost of obtaining further features). In contrast, much of ML is devoted to the unrealistic, sterile environment where all features are observed and further information on an instance is obviated. Here we extend past static ML and develop an active feature acquisition (AFA) framework that interacts with the environment to obtain new information on-the-fly and can: 1) make inferences on an instance in the face of incomplete features, 2) determine a plan for feature acquisitions to obtain additional information on the instance at hand. We build our AFA framework on a backbone of understanding the information and conditional dependencies that are present in the data. First, we show how to build generative models that can capture dependencies over arbitrary subsets of features and employ these models for acquisitions in a greedy scheme. After, we show that it is possible to guide the training of RL agents for AFA via side-information and auxiliary rewards stemming from our generative models. We also examine two important factors for deploying AFA models in real-world scenarios, namely interpretability and robustness. Extensive experiments demonstrate the state-of-the-art performance of our AFA framework.
Towards Cost Sensitive Decision Making
Oct 04, 2024



Abstract:Many real-world situations allow for the acquisition of additional relevant information when making decisions with limited or uncertain data. However, traditional RL approaches either require all features to be acquired beforehand (e.g. in a MDP) or regard part of them as missing data that cannot be acquired (e.g. in a POMDP). In this work, we consider RL models that may actively acquire features from the environment to improve the decision quality and certainty, while automatically balancing the cost of feature acquisition process and the reward of task decision process. We propose the Active-Acquisition POMDP and identify two types of the acquisition process for different application domains. In order to assist the agent in the actively-acquired partially-observed environment and alleviate the exploration-exploitation dilemma, we develop a model-based approach, where a deep generative model is utilized to capture the dependencies of the features and impute the unobserved features. The imputations essentially represent the beliefs of the agent. Equipped with the dynamics model, we develop hierarchical RL algorithms to resolve both types of the AA-POMDPs. Empirical results demonstrate that our approach achieves considerably better performance than existing POMDP-RL solutions.
Localizing Anomalies via Multiscale Score Matching Analysis
Jun 28, 2024


Abstract:Anomaly detection and localization in medical imaging remain critical challenges in healthcare. This paper introduces Spatial-MSMA (Multiscale Score Matching Analysis), a novel unsupervised method for anomaly localization in volumetric brain MRIs. Building upon the MSMA framework, our approach incorporates spatial information and conditional likelihoods to enhance anomaly detection capabilities. We employ a flexible normalizing flow model conditioned on patch positions and global image features to estimate patch-wise anomaly scores. The method is evaluated on a dataset of 1,650 T1- and T2-weighted brain MRIs from typically developing children, with simulated lesions added to the test set. Spatial-MSMA significantly outperforms existing methods, including reconstruction-based, generative-based, and interpretation-based approaches, in lesion detection and segmentation tasks. Our model achieves superior performance in both distance-based metrics (99th percentile Hausdorff Distance: $7.05 \pm 0.61$, Mean Surface Distance: $2.10 \pm 0.43$) and component-wise metrics (True Positive Rate: $0.83 \pm 0.01$, Positive Predictive Value: $0.96 \pm 0.01$). These results demonstrate Spatial-MSMA's potential for accurate and interpretable anomaly localization in medical imaging, with implications for improved diagnosis and treatment planning in clinical settings. Our code is available at~\url{https://github.com/ahsanMah/sade/}.
EMOE: Expansive Matching of Experts for Robust Uncertainty Based Rejection
Jun 03, 2024



Abstract:Expansive Matching of Experts (EMOE) is a novel method that utilizes support-expanding, extrapolatory pseudo-labeling to improve prediction and uncertainty based rejection on out-of-distribution (OOD) points. We propose an expansive data augmentation technique that generates OOD instances in a latent space, and an empirical trial based approach to filter out augmented expansive points for pseudo-labeling. EMOE utilizes a diverse set of multiple base experts as pseudo-labelers on the augmented data to improve OOD performance through a shared MLP with multiple heads (one per expert). We demonstrate that EMOE achieves superior performance compared to state-of-the-art methods on tabular data.
A Unified Model for Longitudinal Multi-Modal Multi-View Prediction with Missingness
Mar 22, 2024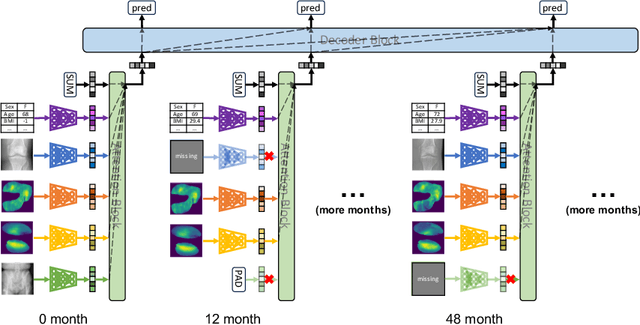
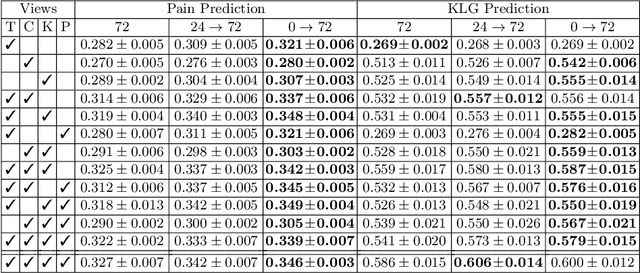
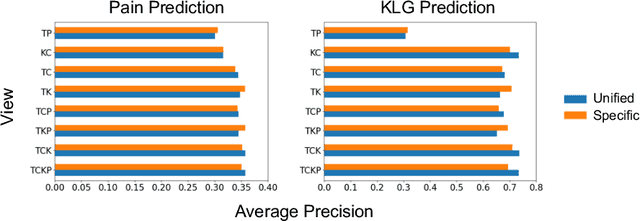
Abstract:Medical records often consist of different modalities, such as images, text, and tabular information. Integrating all modalities offers a holistic view of a patient's condition, while analyzing them longitudinally provides a better understanding of disease progression. However, real-world longitudinal medical records present challenges: 1) patients may lack some or all of the data for a specific timepoint, and 2) certain modalities or views might be absent for all patients during a particular period. In this work, we introduce a unified model for longitudinal multi-modal multi-view prediction with missingness. Our method allows as many timepoints as desired for input, and aims to leverage all available data, regardless of their availability. We conduct extensive experiments on the knee osteoarthritis dataset from the Osteoarthritis Initiative for pain and Kellgren-Lawrence grade prediction at a future timepoint. We demonstrate the effectiveness of our method by comparing results from our unified model to specific models that use the same modality and view combinations during training and evaluation. We also show the benefit of having extended temporal data and provide post-hoc analysis for a deeper understanding of each modality/view's importance for different tasks.
Anomaly Detection via Gumbel Noise Score Matching
Apr 06, 2023



Abstract:We propose Gumbel Noise Score Matching (GNSM), a novel unsupervised method to detect anomalies in categorical data. GNSM accomplishes this by estimating the scores, i.e. the gradients of log likelihoods w.r.t.~inputs, of continuously relaxed categorical distributions. We test our method on a suite of anomaly detection tabular datasets. GNSM achieves a consistently high performance across all experiments. We further demonstrate the flexibility of GNSM by applying it to image data where the model is tasked to detect poor segmentation predictions. Images ranked anomalous by GNSM show clear segmentation failures, with the outputs of GNSM strongly correlating with segmentation metrics computed on ground-truth. We outline the score matching training objective utilized by GNSM and provide an open-source implementation of our work.
Acquisition Conditioned Oracle for Nongreedy Active Feature Acquisition
Feb 27, 2023Abstract:We develop novel methodology for active feature acquisition (AFA), the study of how to sequentially acquire a dynamic (on a per instance basis) subset of features that minimizes acquisition costs whilst still yielding accurate predictions. The AFA framework can be useful in a myriad of domains, including health care applications where the cost of acquiring additional features for a patient (in terms of time, money, risk, etc.) can be weighed against the expected improvement to diagnostic performance. Previous approaches for AFA have employed either: deep learning RL techniques, which have difficulty training policies in the AFA MDP due to sparse rewards and a complicated action space; deep learning surrogate generative models, which require modeling complicated multidimensional conditional distributions; or greedy policies, which fail to account for how joint feature acquisitions can be informative together for better predictions. In this work we show that we can bypass many of these challenges with a novel, nonparametric oracle based approach, which we coin the acquisition conditioned oracle (ACO). Extensive experiments show the superiority of the ACO to state-of-the-art AFA methods when acquiring features for both predictions and general decision-making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge