Jungwon Choi
ReaMIL: Reasoning- and Evidence-Aware Multiple Instance Learning for Whole-Slide Histopathology
Jan 15, 2026Abstract:We introduce ReaMIL (Reasoning- and Evidence-Aware MIL), a multiple instance learning approach for whole-slide histopathology that adds a light selection head to a strong MIL backbone. The head produces soft per-tile gates and is trained with a budgeted-sufficiency objective: a hinge loss that enforces the true-class probability to be $\geq τ$ using only the kept evidence, under a sparsity budget on the number of selected tiles. The budgeted-sufficiency objective yields small, spatially compact evidence sets without sacrificing baseline performance. Across TCGA-NSCLC (LUAD vs. LUSC), TCGA-BRCA (IDC vs. Others), and PANDA, ReaMIL matches or slightly improves baseline AUC and provides quantitative evidence-efficiency diagnostics. On NSCLC, it attains AUC 0.983 with a mean minimal sufficient K (MSK) $\approx 8.2$ tiles at $τ= 0.90$ and AUKC $\approx 0.864$, showing that class confidence rises sharply and stabilizes once a small set of tiles is kept. The method requires no extra supervision, integrates seamlessly with standard MIL training, and naturally yields slide-level overlays. We report accuracy alongside MSK, AUKC, and contiguity for rigorous evaluation of model behavior on WSIs.
A Foundational Brain Dynamics Model via Stochastic Optimal Control
Feb 07, 2025Abstract:We introduce a foundational model for brain dynamics that utilizes stochastic optimal control (SOC) and amortized inference. Our method features a continuous-discrete state space model (SSM) that can robustly handle the intricate and noisy nature of fMRI signals. To address computational limitations, we implement an approximation strategy grounded in the SOC framework. Additionally, we present a simulation-free latent dynamics approach that employs locally linear approximations, facilitating efficient and scalable inference. For effective representation learning, we derive an Evidence Lower Bound (ELBO) from the SOC formulation, which integrates smoothly with recent advancements in self-supervised learning (SSL), thereby promoting robust and transferable representations. Pre-trained on extensive datasets such as the UKB, our model attains state-of-the-art results across a variety of downstream tasks, including demographic prediction, trait analysis, disease diagnosis, and prognosis. Moreover, evaluating on external datasets such as HCP-A, ABIDE, and ADHD200 further validates its superior abilities and resilience across different demographic and clinical distributions. Our foundational model provides a scalable and efficient approach for deciphering brain dynamics, opening up numerous applications in neuroscience.
Stochastic Optimal Control for Diffusion Bridges in Function Spaces
Jun 03, 2024
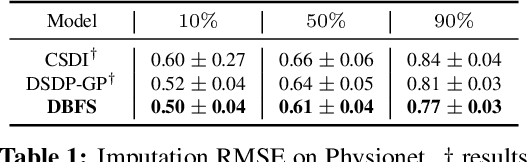
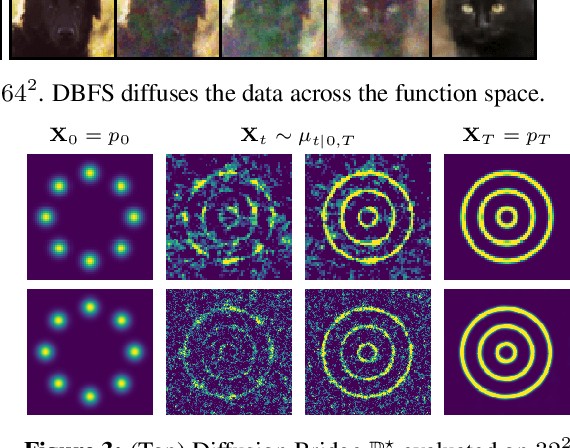
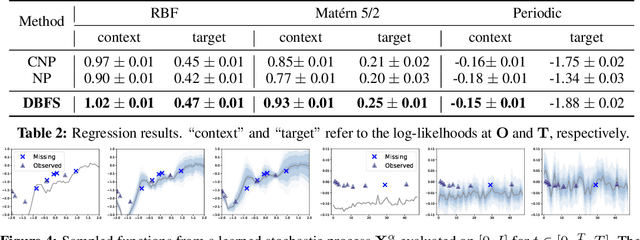
Abstract:Recent advancements in diffusion models and diffusion bridges primarily focus on finite-dimensional spaces, yet many real-world problems necessitate operations in infinite-dimensional function spaces for more natural and interpretable formulations. In this paper, we present a theory of stochastic optimal control (SOC) tailored to infinite-dimensional spaces, aiming to extend diffusion-based algorithms to function spaces. Specifically, we demonstrate how Doob's $h$-transform, the fundamental tool for constructing diffusion bridges, can be derived from the SOC perspective and expanded to infinite dimensions. This expansion presents a challenge, as infinite-dimensional spaces typically lack closed-form densities. Leveraging our theory, we establish that solving the optimal control problem with a specific objective function choice is equivalent to learning diffusion-based generative models. We propose two applications: (1) learning bridges between two infinite-dimensional distributions and (2) generative models for sampling from an infinite-dimensional distribution. Our approach proves effective for diverse problems involving continuous function space representations, such as resolution-free images, time-series data, and probability density functions.
Joint-Embedding Masked Autoencoder for Self-supervised Learning of Dynamic Functional Connectivity from the Human Brain
Mar 11, 2024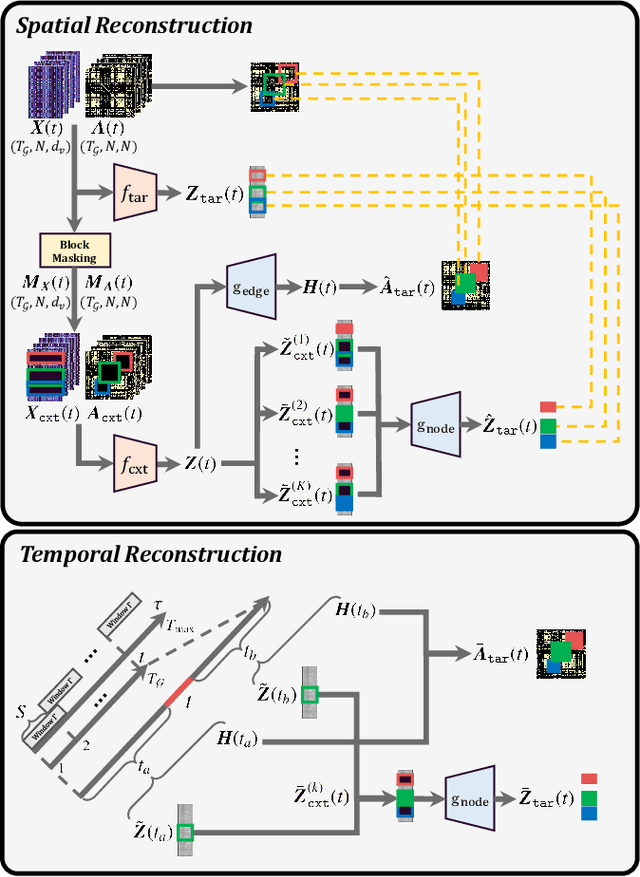
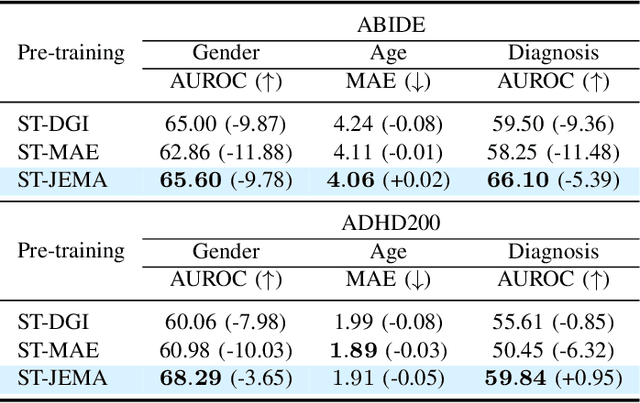
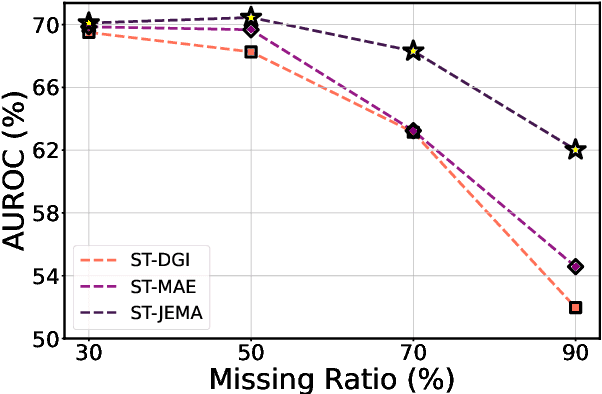
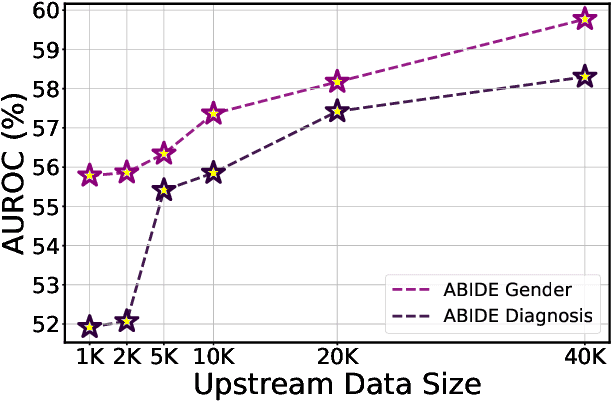
Abstract:Graph Neural Networks (GNNs) have shown promise in learning dynamic functional connectivity for distinguishing phenotypes from human brain networks. However, obtaining extensive labeled clinical data for training is often resource-intensive, making practical application difficult. Leveraging unlabeled data thus becomes crucial for representation learning in a label-scarce setting. Although generative self-supervised learning techniques, especially masked autoencoders, have shown promising results in representation learning in various domains, their application to dynamic graphs for dynamic functional connectivity remains underexplored, facing challenges in capturing high-level semantic representations. Here, we introduce the Spatio-Temporal Joint Embedding Masked Autoencoder (ST-JEMA), drawing inspiration from the Joint Embedding Predictive Architecture (JEPA) in computer vision. ST-JEMA employs a JEPA-inspired strategy for reconstructing dynamic graphs, which enables the learning of higher-level semantic representations considering temporal perspectives, addressing the challenges in fMRI data representation learning. Utilizing the large-scale UK Biobank dataset for self-supervised learning, ST-JEMA shows exceptional representation learning performance on dynamic functional connectivity demonstrating superiority over previous methods in predicting phenotypes and psychiatric diagnoses across eight benchmark fMRI datasets even with limited samples and effectiveness of temporal reconstruction on missing data scenarios. These findings highlight the potential of our approach as a robust representation learning method for leveraging label-scarce fMRI data.
A Generative Self-Supervised Framework using Functional Connectivity in fMRI Data
Dec 04, 2023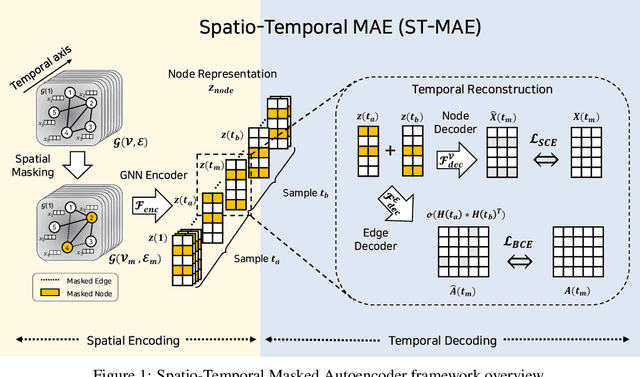
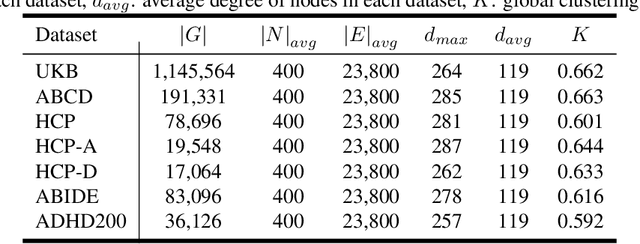
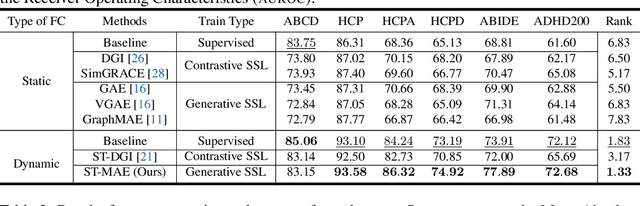
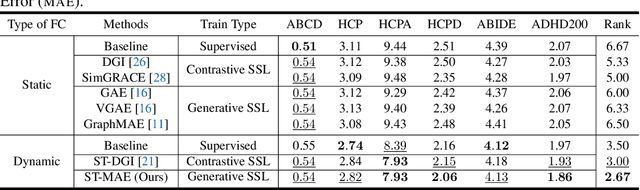
Abstract:Deep neural networks trained on Functional Connectivity (FC) networks extracted from functional Magnetic Resonance Imaging (fMRI) data have gained popularity due to the increasing availability of data and advances in model architectures, including Graph Neural Network (GNN). Recent research on the application of GNN to FC suggests that exploiting the time-varying properties of the FC could significantly improve the accuracy and interpretability of the model prediction. However, the high cost of acquiring high-quality fMRI data and corresponding phenotypic labels poses a hurdle to their application in real-world settings, such that a model na\"ively trained in a supervised fashion can suffer from insufficient performance or a lack of generalization on a small number of data. In addition, most Self-Supervised Learning (SSL) approaches for GNNs to date adopt a contrastive strategy, which tends to lose appropriate semantic information when the graph structure is perturbed or does not leverage both spatial and temporal information simultaneously. In light of these challenges, we propose a generative SSL approach that is tailored to effectively harness spatio-temporal information within dynamic FC. Our empirical results, experimented with large-scale (>50,000) fMRI datasets, demonstrate that our approach learns valuable representations and enables the construction of accurate and robust models when fine-tuned for downstream tasks.
On Divergence Measures for Bayesian Pseudocoresets
Oct 12, 2022



Abstract:A Bayesian pseudocoreset is a small synthetic dataset for which the posterior over parameters approximates that of the original dataset. While promising, the scalability of Bayesian pseudocoresets is not yet validated in realistic problems such as image classification with deep neural networks. On the other hand, dataset distillation methods similarly construct a small dataset such that the optimization using the synthetic dataset converges to a solution with performance competitive with optimization using full data. Although dataset distillation has been empirically verified in large-scale settings, the framework is restricted to point estimates, and their adaptation to Bayesian inference has not been explored. This paper casts two representative dataset distillation algorithms as approximations to methods for constructing pseudocoresets by minimizing specific divergence measures: reverse KL divergence and Wasserstein distance. Furthermore, we provide a unifying view of such divergence measures in Bayesian pseudocoreset construction. Finally, we propose a novel Bayesian pseudocoreset algorithm based on minimizing forward KL divergence. Our empirical results demonstrate that the pseudocoresets constructed from these methods reflect the true posterior even in high-dimensional Bayesian inference problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge