John Gounley
Pixel-Resolved Long-Context Learning for Turbulence at Exascale: Resolving Small-scale Eddies Toward the Viscous Limit
Jul 22, 2025Abstract:Turbulence plays a crucial role in multiphysics applications, including aerodynamics, fusion, and combustion. Accurately capturing turbulence's multiscale characteristics is essential for reliable predictions of multiphysics interactions, but remains a grand challenge even for exascale supercomputers and advanced deep learning models. The extreme-resolution data required to represent turbulence, ranging from billions to trillions of grid points, pose prohibitive computational costs for models based on architectures like vision transformers. To address this challenge, we introduce a multiscale hierarchical Turbulence Transformer that reduces sequence length from billions to a few millions and a novel RingX sequence parallelism approach that enables scalable long-context learning. We perform scaling and science runs on the Frontier supercomputer. Our approach demonstrates excellent performance up to 1.1 EFLOPS on 32,768 AMD GPUs, with a scaling efficiency of 94%. To our knowledge, this is the first AI model for turbulence that can capture small-scale eddies down to the dissipative range.
Learning to Diagnose Privately: DP-Powered LLMs for Radiology Report Classification
Jun 04, 2025Abstract:Purpose: This study proposes a framework for fine-tuning large language models (LLMs) with differential privacy (DP) to perform multi-abnormality classification on radiology report text. By injecting calibrated noise during fine-tuning, the framework seeks to mitigate the privacy risks associated with sensitive patient data and protect against data leakage while maintaining classification performance. Materials and Methods: We used 50,232 radiology reports from the publicly available MIMIC-CXR chest radiography and CT-RATE computed tomography datasets, collected between 2011 and 2019. Fine-tuning of LLMs was conducted to classify 14 labels from MIMIC-CXR dataset, and 18 labels from CT-RATE dataset using Differentially Private Low-Rank Adaptation (DP-LoRA) in high and moderate privacy regimes (across a range of privacy budgets = {0.01, 0.1, 1.0, 10.0}). Model performance was evaluated using weighted F1 score across three model architectures: BERT-medium, BERT-small, and ALBERT-base. Statistical analyses compared model performance across different privacy levels to quantify the privacy-utility trade-off. Results: We observe a clear privacy-utility trade-off through our experiments on 2 different datasets and 3 different models. Under moderate privacy guarantees the DP fine-tuned models achieved comparable weighted F1 scores of 0.88 on MIMIC-CXR and 0.59 on CT-RATE, compared to non-private LoRA baselines of 0.90 and 0.78, respectively. Conclusion: Differentially private fine-tuning using LoRA enables effective and privacy-preserving multi-abnormality classification from radiology reports, addressing a key challenge in fine-tuning LLMs on sensitive medical data.
MATEY: multiscale adaptive foundation models for spatiotemporal physical systems
Dec 29, 2024
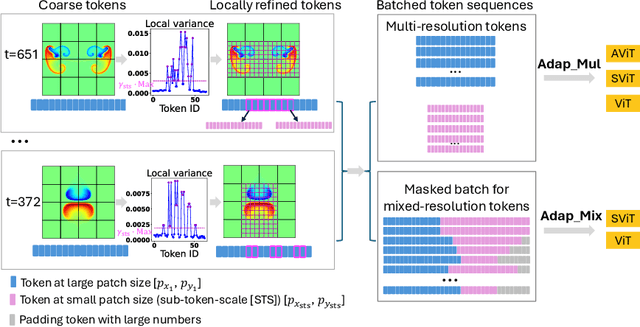


Abstract:Accurate representation of the multiscale features in spatiotemporal physical systems using vision transformer (ViT) architectures requires extremely long, computationally prohibitive token sequences. To address this issue, we propose two adaptive tokenization schemes that dynamically adjust patch sizes based on local features: one ensures convergent behavior to uniform patch refinement, while the other offers better computational efficiency. Moreover, we present a set of spatiotemporal attention schemes, where the temporal or axial spatial dimensions are decoupled, and evaluate their computational and data efficiencies. We assess the performance of the proposed multiscale adaptive model, MATEY, in a sequence of experiments. The results show that adaptive tokenization schemes achieve improved accuracy without significantly increasing the length of the token sequence. Compared to a full spatiotemporal attention scheme or a scheme that decouples only the temporal dimension, we find that fully decoupled axial attention is less efficient and expressive, requiring more training time and model weights to achieve the same accuracy. Finally, we demonstrate in two fine-tuning tasks featuring different physics that models pretrained on PDEBench data outperform the ones trained from scratch, especially in the low data regime with frozen attention.
Adiabatic Quantum Support Vector Machines
Jan 23, 2024Abstract:Adiabatic quantum computers can solve difficult optimization problems (e.g., the quadratic unconstrained binary optimization problem), and they seem well suited to train machine learning models. In this paper, we describe an adiabatic quantum approach for training support vector machines. We show that the time complexity of our quantum approach is an order of magnitude better than the classical approach. Next, we compare the test accuracy of our quantum approach against a classical approach that uses the Scikit-learn library in Python across five benchmark datasets (Iris, Wisconsin Breast Cancer (WBC), Wine, Digits, and Lambeq). We show that our quantum approach obtains accuracies on par with the classical approach. Finally, we perform a scalability study in which we compute the total training times of the quantum approach and the classical approach with increasing number of features and number of data points in the training dataset. Our scalability results show that the quantum approach obtains a 3.5--4.5 times speedup over the classical approach on datasets with many (millions of) features.
Transferring a molecular foundation model for polymer property predictions
Oct 25, 2023Abstract:Transformer-based large language models have remarkable potential to accelerate design optimization for applications such as drug development and materials discovery. Self-supervised pretraining of transformer models requires large-scale datasets, which are often sparsely populated in topical areas such as polymer science. State-of-the-art approaches for polymers conduct data augmentation to generate additional samples but unavoidably incurs extra computational costs. In contrast, large-scale open-source datasets are available for small molecules and provide a potential solution to data scarcity through transfer learning. In this work, we show that using transformers pretrained on small molecules and fine-tuned on polymer properties achieve comparable accuracy to those trained on augmented polymer datasets for a series of benchmark prediction tasks.
DeepSpeed4Science Initiative: Enabling Large-Scale Scientific Discovery through Sophisticated AI System Technologies
Oct 11, 2023



Abstract:In the upcoming decade, deep learning may revolutionize the natural sciences, enhancing our capacity to model and predict natural occurrences. This could herald a new era of scientific exploration, bringing significant advancements across sectors from drug development to renewable energy. To answer this call, we present DeepSpeed4Science initiative (deepspeed4science.ai) which aims to build unique capabilities through AI system technology innovations to help domain experts to unlock today's biggest science mysteries. By leveraging DeepSpeed's current technology pillars (training, inference and compression) as base technology enablers, DeepSpeed4Science will create a new set of AI system technologies tailored for accelerating scientific discoveries by addressing their unique complexity beyond the common technical approaches used for accelerating generic large language models (LLMs). In this paper, we showcase the early progress we made with DeepSpeed4Science in addressing two of the critical system challenges in structural biology research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge