Jitender Saini
Graph Classification and Radiomics Signature for Identification of Tuberculous Meningitis
Apr 01, 2025
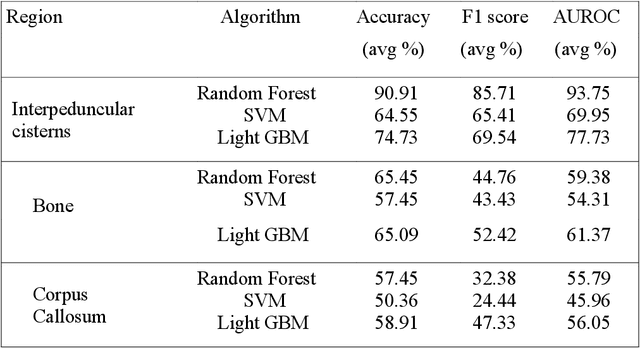


Abstract:Introduction: Tuberculous meningitis (TBM) is a serious brain infection caused by Mycobacterium tuberculosis, characterized by inflammation of the meninges covering the brain and spinal cord. Diagnosis often requires invasive lumbar puncture (LP) and cerebrospinal fluid (CSF) analysis. Objectives: This study aims to classify TBM patients using T1-weighted (T1w) non-contrast Magnetic Resonance Imaging (MRI) scans. We hypothesize that specific brain regions, such as the interpeduncular cisterns, bone, and corpus callosum, contain visual markers that can non-invasively distinguish TBM patients from healthy controls. We propose a novel Pixel-array Graphs Classifier (PAG-Classifier) that leverages spatial relationships between neighbouring 3D pixels in a graph-based framework to extract significant features through eigen decomposition. These features are then used to train machine learning classifiers for effective patient classification. We validate our approach using a radiomics-based methodology, classifying TBM patients based on relevant radiomics features. Results: We utilized an internal dataset consisting of 52 scans, 32 from confirmed TBM patients based on mycobacteria detection in CSF, and 20 from healthy individuals. We achieved a 5-fold cross-validated average F1 score of 85.71% for cistern regions with our PAG-Classifier and 92.85% with the radiomics features classifier, surpassing current state-of-the-art benchmarks by 15% and 22%, respectively. However, bone and corpus callosum regions showed poor classification effectiveness, with average F1 scores below 50%. Conclusion: Our study suggests that algorithms like the PAG-Classifier serve as effective tools for non-invasive TBM analysis, particularly by targeting the interpeduncular cistern. Findings indicate that the bone and corpus callosum regions lack distinctive patterns for differentiation.
Labeling subtypes in a Parkinson's Cohort using Multifeatures in MRI -- Integrating Grey and White Matter Information
Mar 26, 2024



Abstract:Thresholding of networks has long posed a challenge in brain connectivity analysis. Weighted networks are typically binarized using threshold measures to facilitate network analysis. Previous studies on MRI-based brain networks have predominantly utilized density or sparsity-based thresholding techniques, optimized within specific ranges derived from network metrics such as path length, clustering coefficient, and small-world index. Thus, determination of a single threshold value for facilitating comparative analysis of networks remains elusive. To address this, our study introduces Mutual K-Nearest Neighbor (MKNN)-based thresholding for brain network analysis. Here, nearest neighbor selection is based on the highest correlation between features of brain regions. Construction of brain networks was accomplished by computing Pearson correlations between grey matter volume and white matter volume for each pair of brain regions. Structural MRI data from 180 Parkinsons patients and 70 controls from the NIMHANS, India were analyzed. Subtypes within Parkinsons disease were identified based on grey and white matter volume atrophy using source-based morphometric decomposition. The loading coefficients were correlated with clinical features to discern clinical relationship with the deciphered subtypes. Our data-mining approach revealed: Subtype A (N = 51, intermediate type), Subtype B (N = 57, mild-severe type with mild motor symptoms), and Subtype AB (N = 36, most-severe type with predominance in motor impairment). Subtype-specific weighted matrices were binarized using MKNN-based thresholding for brain network analysis. Permutation tests on network metrics of resulting bipartite graphs demonstrated significant group differences in betweenness centrality and participation coefficient. The identified hubs were specific to each subtype, with some hubs conserved across different subtypes.
Unique Brain Network Identification Number for Parkinson's Individuals Using Structural MRI
Jun 02, 2023Abstract:We propose a novel algorithm called Unique Brain Network Identification Number (UBNIN) for encoding brain networks of individual subject. To realize this objective, we employed T1-weighted structural MRI of 180 Parkinson's disease (PD) patients from National Institute of Mental Health and Neurosciences, India. We parcellated each subject's brain volume and constructed individual adjacency matrix using correlation between grey matter (GM) volume of every pair of regions. The unique code is derived from values representing connections of every node (i), weighted by a factor of 2^-(i-1). The numerical representation UBNIN was observed to be distinct for each individual brain network, which may also be applied to other neuroimaging modalities. This model may be implemented as neural signature of a person's unique brain connectivity, thereby useful for brainprinting applications. Additionally, we segregated the above dataset into five age-cohorts: A:22-32years, B:33-42years, C:43-52years, D:53-62years and E:63-72years to study the variation in network topology over age. Sparsity was adopted as the threshold estimate to binarize each age-based correlation matrix. Connectivity metrics were obtained using Brain Connectivity toolbox-based MATLAB functions. For each age-cohort, a decreasing trend was observed in mean clustering coefficient with increasing sparsity. Significantly different clustering coefficient was noted between age-cohort B and C (sparsity: 0.63,0.66), C and E (sparsity: 0.66,0.69). Our findings suggest network connectivity patterns change with age, indicating network disruption due to the underlying neuropathology. Varying clustering coefficient for different cohorts indicate that information transfer between neighboring nodes change with age. This provides evidence on age-related brain shrinkage and network degeneration.
Interpretable simultaneous localization of MRI corpus callosum and classification of atypical Parkinsonian disorders using YOLOv5
Jun 01, 2023Abstract:Structural MRI(S-MRI) is one of the most versatile imaging modality that revolutionized the anatomical study of brain in past decades. The corpus callosum (CC) is the principal white matter fibre tract, enabling all kinds of inter-hemispheric communication. Thus, subtle changes in CC might be associated with various neurological disorders. The present work proposes the potential of YOLOv5-based CC detection framework to differentiate atypical Parkinsonian disorders (PD) from healthy controls (HC). With 3 rounds of hold-out validation, mean classification accuracy of 92% is obtained using the proposed method on a proprietary dataset consisting of 20 healthy subjects and 20 cases of APDs, with an improvement of 5% over SOTA methods (CC morphometry and visual texture analysis) that used the same dataset. Subsequently, in order to incorporate the explainability of YOLO predictions, Eigen CAM based heatmap is generated for identifying the most important sub-region in CC that leads to the classification. The result of Eigen CAM showed CC mid-body as the most distinguishable sub-region in classifying APDs and HC, which is in-line with SOTA methodologies and the current prevalent understanding in medicine.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
HR-CAM: Precise Localization of Pathology Using Multi-level Learning in CNNs
Sep 23, 2019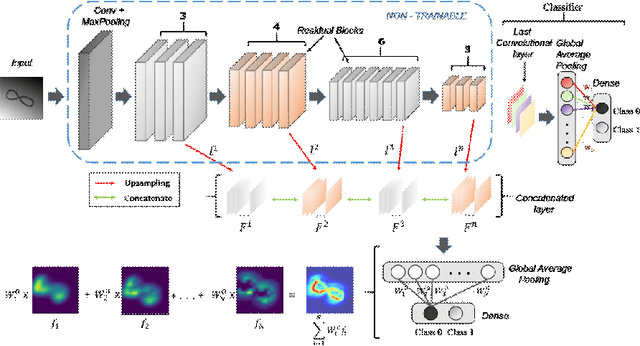
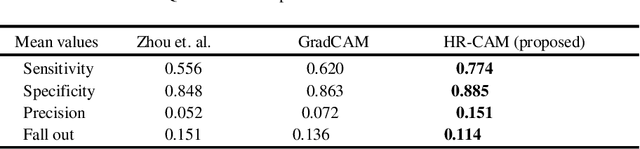
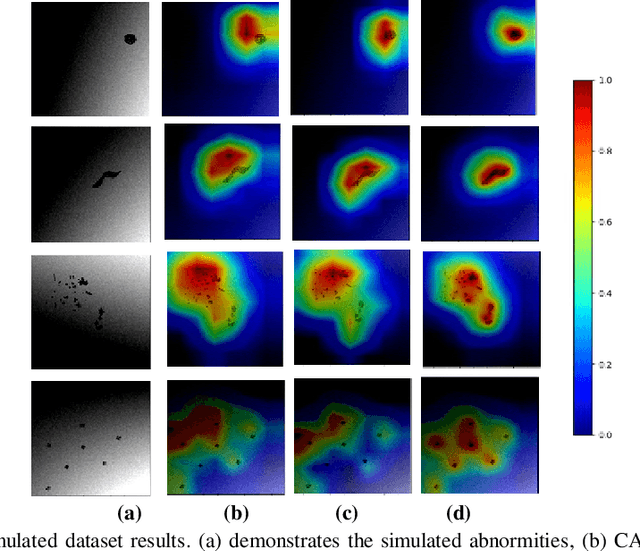
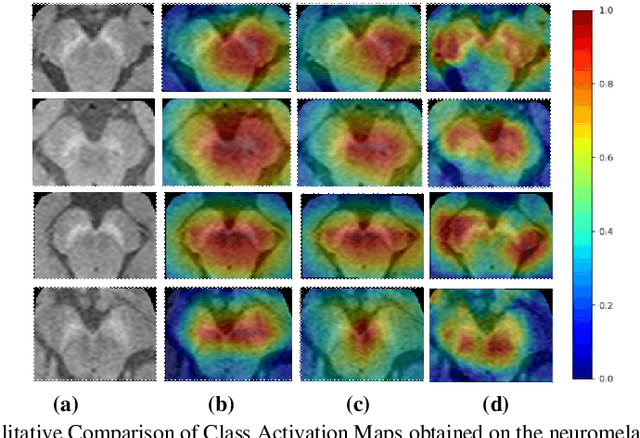
Abstract:We propose a CNN based technique that aggregates feature maps from its multiple layers that can localize abnormalities with greater details as well as predict pathology under consideration. Existing class activation mapping (CAM) techniques extract feature maps from either the final layer or a single intermediate layer to create the discriminative maps and then interpolate to upsample to the original image resolution. In this case, the subject specific localization is coarse and is unable to capture subtle abnormalities. To mitigate this, our method builds a novel CNN based discriminative localization model that we call high resolution CAM (HR-CAM), which accounts for layers from each resolution, therefore facilitating a comprehensive map that can delineate the pathology for each subject by combining low-level, intermediate as well as high-level features from the CNN. Moreover, our model directly provides the discriminative map in the resolution of the original image facilitating finer delineation of abnormalities. We demonstrate the working of our model on a simulated abnormalities data where we illustrate how the model captures finer details in the final discriminative maps as compared to current techniques. We then apply this technique: (1) to classify ependymomas from grade IV glioblastoma on T1-weighted contrast enhanced (T1-CE) MRI and (2) to predict Parkinson's disease from neuromelanin sensitive MRI. In all these cases we demonstrate that our model not only predicts pathologies with high accuracies, but also creates clinically interpretable subject specific high resolution discriminative localizations. Overall, the technique can be generalized to any CNN and carries high relevance in a clinical setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge