Cota Navin Gupta
Dynamical Embedding of Single Channel Electroencephalogram for Artifact Subspace Reconstruction
Jun 28, 2024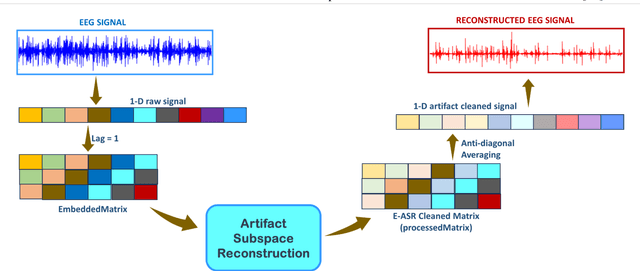
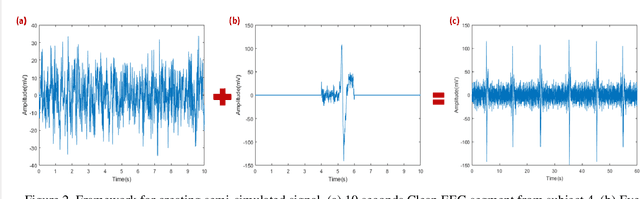
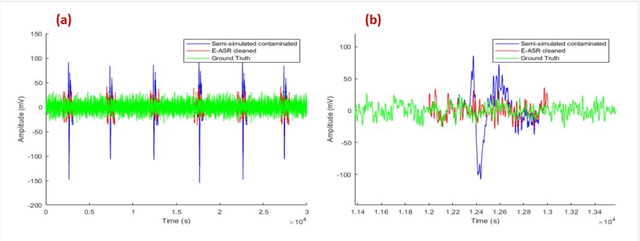
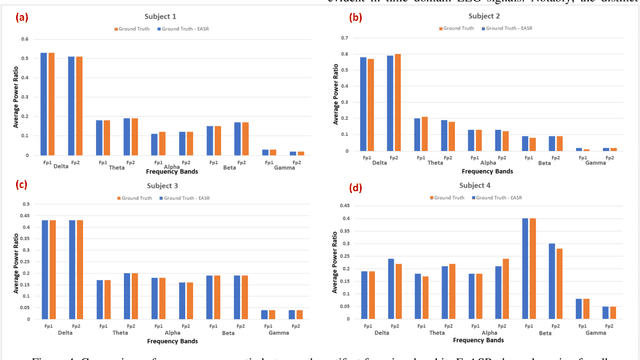
Abstract:This study introduces a novel framework to apply Artifact Subspace Reconstruction (ASR) algorithm on single-channel Electroencephalogram (EEG) data. ASR, renowned for its automated capability to effectively eliminate various artifacts like eye-blinks and eye movements from EEG signals. Importantly it has been implemented on android smartphones, but relied on multiple channels for principal component subspace calculations. To overcome this limitation, we incorporate the established dynamical embedding approach into the algorithm, naming it Embedded-ASR (E-ASR). In our proposed method, an embedded matrix is first constructed from a single-channel EEG data using series of delay vectors. ASR is then applied to this embedded matrix, and the resulting cleaned single-channel EEG is reconstructed by removing the time lag and concatenating the rows of the embedded matrix. Data was collected from four subjects in resting states with eyes open from pre-frontal (Fp1 and Fp2) electrodes using CameraEEG app. To assess the effectiveness of the E-ASR algorithm on an EEG dataset with artifacts, we employed performance metrics such as relative root mean square error (RRMSE), correlation coefficient (CC), average power ratio as well as estimated the number of eye-blinks with and without the E-ASR approach. E-ASR was able to reduce artifacts from the semi-simulated EEG data, with an RRMSE of 45.45% and a CC of 0.91. For real EEG data, the counted eye-blinks were manually cross-checked with ground truth obtained from CameraEEG video data across all subjects for individual Fp1 and Fp2 electrodes. In conclusion, our study suggests E-ASR framework can remove artifacts from single channel EEG data. This promising algorithm might have potential for smartphone-based natural environment EEG applications, where minimal number of electrodes is a critical factor.
Systematic Review of Experimental Paradigms and Deep Neural Networks for Electroencephalography-Based Cognitive Workload Detection
Sep 11, 2023Abstract:This article summarizes a systematic review of the electroencephalography (EEG)-based cognitive workload (CWL) estimation. The focus of the article is twofold: identify the disparate experimental paradigms used for reliably eliciting discreet and quantifiable levels of cognitive load and the specific nature and representational structure of the commonly used input formulations in deep neural networks (DNNs) used for signal classification. The analysis revealed a number of studies using EEG signals in its native representation of a two-dimensional matrix for offline classification of CWL. However, only a few studies adopted an online or pseudo-online classification strategy for real-time CWL estimation. Further, only a couple of interpretable DNNs and a single generative model were employed for cognitive load detection till date during this review. More often than not, researchers were using DNNs as black-box type models. In conclusion, DNNs prove to be valuable tools for classifying EEG signals, primarily due to the substantial modeling power provided by the depth of their network architecture. It is further suggested that interpretable and explainable DNN models must be employed for cognitive workload estimation since existing methods are limited in the face of the non-stationary nature of the signal.
Unique Brain Network Identification Number for Parkinson's Individuals Using Structural MRI
Jun 02, 2023Abstract:We propose a novel algorithm called Unique Brain Network Identification Number (UBNIN) for encoding brain networks of individual subject. To realize this objective, we employed T1-weighted structural MRI of 180 Parkinson's disease (PD) patients from National Institute of Mental Health and Neurosciences, India. We parcellated each subject's brain volume and constructed individual adjacency matrix using correlation between grey matter (GM) volume of every pair of regions. The unique code is derived from values representing connections of every node (i), weighted by a factor of 2^-(i-1). The numerical representation UBNIN was observed to be distinct for each individual brain network, which may also be applied to other neuroimaging modalities. This model may be implemented as neural signature of a person's unique brain connectivity, thereby useful for brainprinting applications. Additionally, we segregated the above dataset into five age-cohorts: A:22-32years, B:33-42years, C:43-52years, D:53-62years and E:63-72years to study the variation in network topology over age. Sparsity was adopted as the threshold estimate to binarize each age-based correlation matrix. Connectivity metrics were obtained using Brain Connectivity toolbox-based MATLAB functions. For each age-cohort, a decreasing trend was observed in mean clustering coefficient with increasing sparsity. Significantly different clustering coefficient was noted between age-cohort B and C (sparsity: 0.63,0.66), C and E (sparsity: 0.66,0.69). Our findings suggest network connectivity patterns change with age, indicating network disruption due to the underlying neuropathology. Varying clustering coefficient for different cohorts indicate that information transfer between neighboring nodes change with age. This provides evidence on age-related brain shrinkage and network degeneration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge