Madhura Ingalhalikar
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
HR-CAM: Precise Localization of Pathology Using Multi-level Learning in CNNs
Sep 23, 2019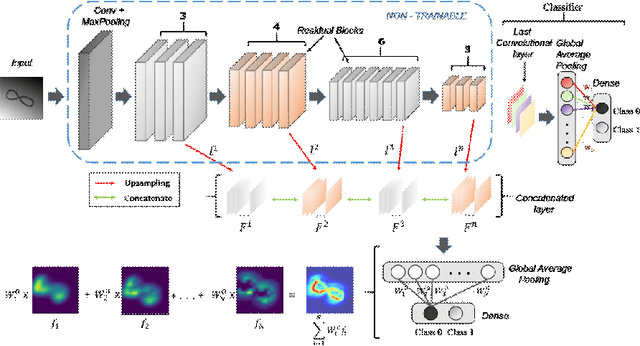
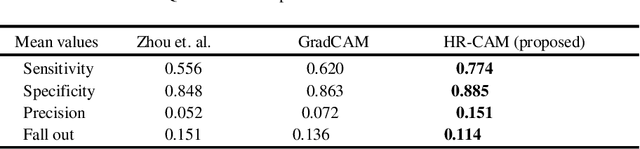
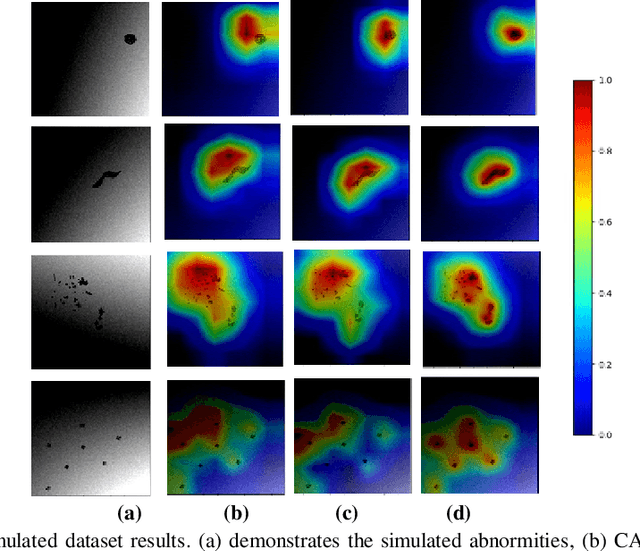
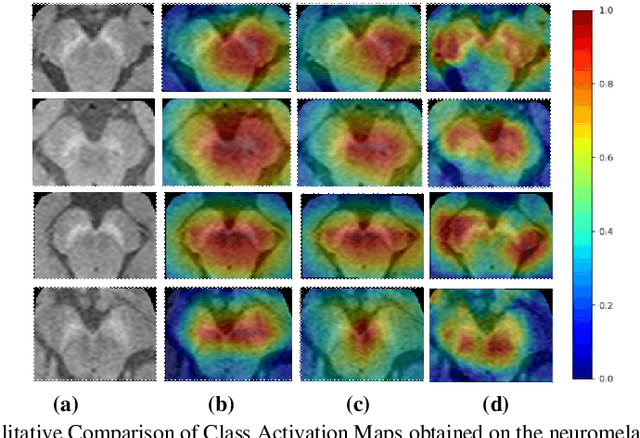
Abstract:We propose a CNN based technique that aggregates feature maps from its multiple layers that can localize abnormalities with greater details as well as predict pathology under consideration. Existing class activation mapping (CAM) techniques extract feature maps from either the final layer or a single intermediate layer to create the discriminative maps and then interpolate to upsample to the original image resolution. In this case, the subject specific localization is coarse and is unable to capture subtle abnormalities. To mitigate this, our method builds a novel CNN based discriminative localization model that we call high resolution CAM (HR-CAM), which accounts for layers from each resolution, therefore facilitating a comprehensive map that can delineate the pathology for each subject by combining low-level, intermediate as well as high-level features from the CNN. Moreover, our model directly provides the discriminative map in the resolution of the original image facilitating finer delineation of abnormalities. We demonstrate the working of our model on a simulated abnormalities data where we illustrate how the model captures finer details in the final discriminative maps as compared to current techniques. We then apply this technique: (1) to classify ependymomas from grade IV glioblastoma on T1-weighted contrast enhanced (T1-CE) MRI and (2) to predict Parkinson's disease from neuromelanin sensitive MRI. In all these cases we demonstrate that our model not only predicts pathologies with high accuracies, but also creates clinically interpretable subject specific high resolution discriminative localizations. Overall, the technique can be generalized to any CNN and carries high relevance in a clinical setting.
ARiA: Utilizing Richard's Curve for Controlling the Non-monotonicity of the Activation Function in Deep Neural Nets
May 22, 2018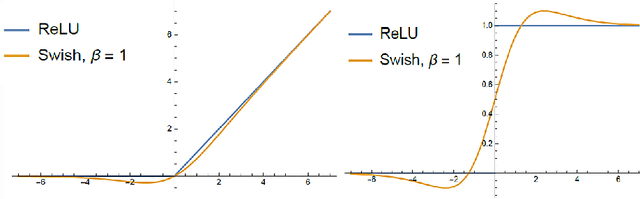
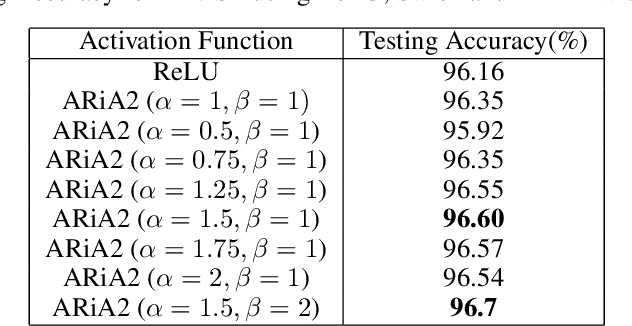
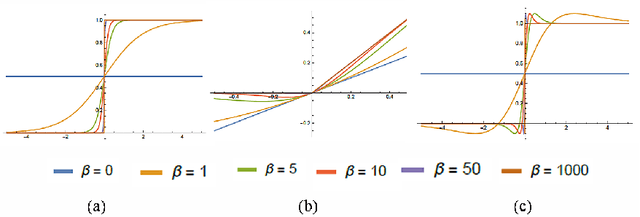

Abstract:This work introduces a novel activation unit that can be efficiently employed in deep neural nets (DNNs) and performs significantly better than the traditional Rectified Linear Units (ReLU). The function developed is a two parameter version of the specialized Richard's Curve and we call it Adaptive Richard's Curve weighted Activation (ARiA). This function is non-monotonous, analogous to the newly introduced Swish, however allows a precise control over its non-monotonous convexity by varying the hyper-parameters. We first demonstrate the mathematical significance of the two parameter ARiA followed by its application to benchmark problems such as MNIST, CIFAR-10 and CIFAR-100, where we compare the performance with ReLU and Swish units. Our results illustrate a significantly superior performance on all these datasets, making ARiA a potential replacement for ReLU and other activations in DNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge