Jinxin Zhu
Towards Open-World Mobile Manipulation in Homes: Lessons from the Neurips 2023 HomeRobot Open Vocabulary Mobile Manipulation Challenge
Jul 09, 2024



Abstract:In order to develop robots that can effectively serve as versatile and capable home assistants, it is crucial for them to reliably perceive and interact with a wide variety of objects across diverse environments. To this end, we proposed Open Vocabulary Mobile Manipulation as a key benchmark task for robotics: finding any object in a novel environment and placing it on any receptacle surface within that environment. We organized a NeurIPS 2023 competition featuring both simulation and real-world components to evaluate solutions to this task. Our baselines on the most challenging version of this task, using real perception in simulation, achieved only an 0.8% success rate; by the end of the competition, the best participants achieved an 10.8\% success rate, a 13x improvement. We observed that the most successful teams employed a variety of methods, yet two common threads emerged among the best solutions: enhancing error detection and recovery, and improving the integration of perception with decision-making processes. In this paper, we detail the results and methodologies used, both in simulation and real-world settings. We discuss the lessons learned and their implications for future research. Additionally, we compare performance in real and simulated environments, emphasizing the necessity for robust generalization to novel settings.
Label-efficient Multi-organ Segmentation Method with Diffusion Model
Feb 23, 2024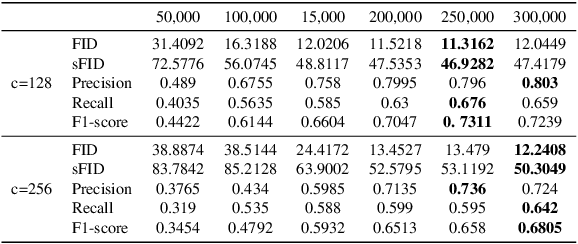
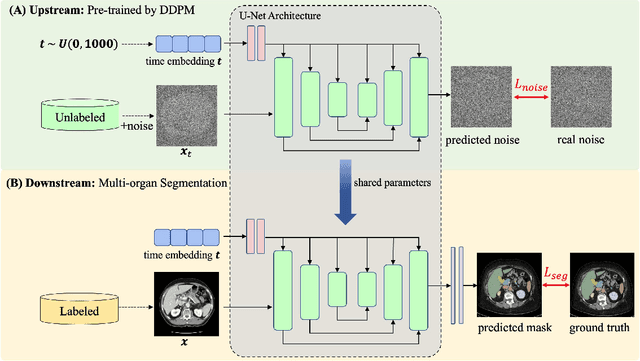

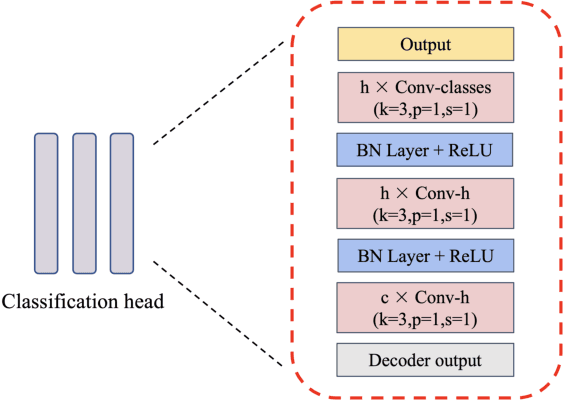
Abstract:Accurate segmentation of multiple organs in Computed Tomography (CT) images plays a vital role in computer-aided diagnosis systems. Various supervised-learning approaches have been proposed recently. However, these methods heavily depend on a large amount of high-quality labeled data, which is expensive to obtain in practice. In this study, we present a label-efficient learning approach using a pre-trained diffusion model for multi-organ segmentation tasks in CT images. First, a denoising diffusion model was trained using unlabeled CT data, generating additional two-dimensional (2D) CT images. Then the pre-trained denoising diffusion network was transferred to the downstream multi-organ segmentation task, effectively creating a semi-supervised learning model that requires only a small amount of labeled data. Furthermore, linear classification and fine-tuning decoder strategies were employed to enhance the network's segmentation performance. Our generative model at 256x256 resolution achieves impressive performance in terms of Fr\'echet inception distance, spatial Fr\'echet inception distance, and F1-score, with values of 11.32, 46.93, and 73.1\%, respectively. These results affirm the diffusion model's ability to generate diverse and realistic 2D CT images. Additionally, our method achieves competitive multi-organ segmentation performance compared to state-of-the-art methods on the FLARE 2022 dataset, particularly in limited labeled data scenarios. Remarkably, even with only 1\% and 10\% labeled data, our method achieves Dice similarity coefficients (DSCs) of 71.56\% and 78.51\% after fine-tuning, respectively. The method achieves a DSC score of 51.81\% using just four labeled CT scans. These results demonstrate the efficacy of our approach in overcoming the limitations of supervised learning heavily reliant on large-scale labeled data.
Ins-ATP: Deep Estimation of ATP for Organoid Based on High Throughput Microscopic Images
Mar 15, 2023



Abstract:Adenosine triphosphate (ATP) is a high-energy phosphate compound and the most direct energy source in organisms. ATP is an essential biomarker for evaluating cell viability in biology. Researchers often use ATP bioluminescence to measure the ATP of organoid after drug to evaluate the drug efficacy. However, ATP bioluminescence has some limitations, leading to unreliable drug screening results. Performing ATP bioluminescence causes cell lysis of organoids, so it is impossible to observe organoids' long-term viability changes after medication continually. To overcome the disadvantages of ATP bioluminescence, we propose Ins-ATP, a non-invasive strategy, the first organoid ATP estimation model based on the high-throughput microscopic image. Ins-ATP directly estimates the ATP of organoids from high-throughput microscopic images, so that it does not influence the drug reactions of organoids. Therefore, the ATP change of organoids can be observed for a long time to obtain more stable results. Experimental results show that the ATP estimation by Ins-ATP is in good agreement with those determined by ATP bioluminescence. Specifically, the predictions of Ins-ATP are consistent with the results measured by ATP bioluminescence in the efficacy evaluation experiments of different drugs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge