Yongzhi Huang
Label-efficient Multi-organ Segmentation Method with Diffusion Model
Feb 23, 2024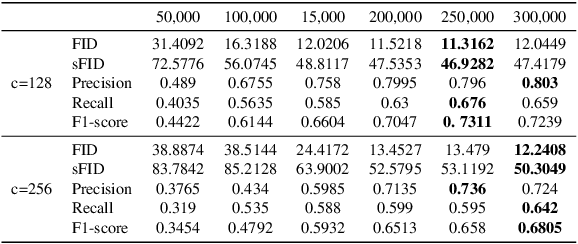
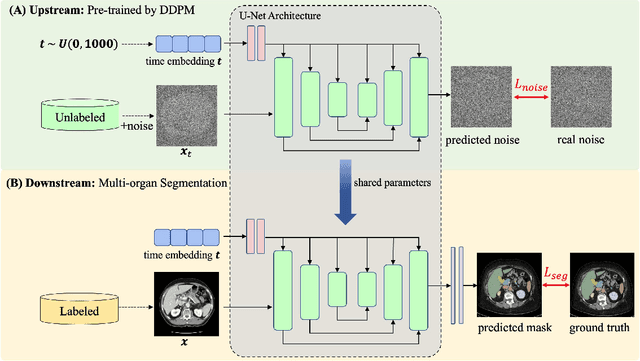

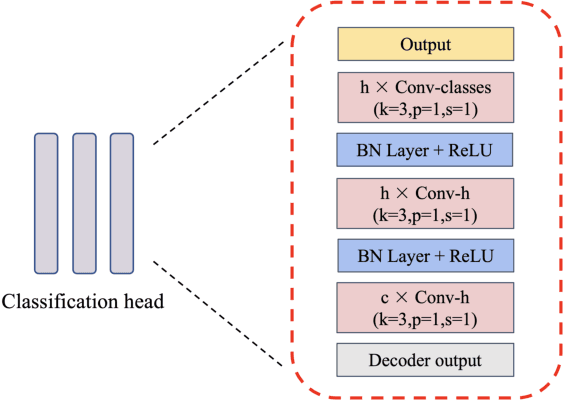
Abstract:Accurate segmentation of multiple organs in Computed Tomography (CT) images plays a vital role in computer-aided diagnosis systems. Various supervised-learning approaches have been proposed recently. However, these methods heavily depend on a large amount of high-quality labeled data, which is expensive to obtain in practice. In this study, we present a label-efficient learning approach using a pre-trained diffusion model for multi-organ segmentation tasks in CT images. First, a denoising diffusion model was trained using unlabeled CT data, generating additional two-dimensional (2D) CT images. Then the pre-trained denoising diffusion network was transferred to the downstream multi-organ segmentation task, effectively creating a semi-supervised learning model that requires only a small amount of labeled data. Furthermore, linear classification and fine-tuning decoder strategies were employed to enhance the network's segmentation performance. Our generative model at 256x256 resolution achieves impressive performance in terms of Fr\'echet inception distance, spatial Fr\'echet inception distance, and F1-score, with values of 11.32, 46.93, and 73.1\%, respectively. These results affirm the diffusion model's ability to generate diverse and realistic 2D CT images. Additionally, our method achieves competitive multi-organ segmentation performance compared to state-of-the-art methods on the FLARE 2022 dataset, particularly in limited labeled data scenarios. Remarkably, even with only 1\% and 10\% labeled data, our method achieves Dice similarity coefficients (DSCs) of 71.56\% and 78.51\% after fine-tuning, respectively. The method achieves a DSC score of 51.81\% using just four labeled CT scans. These results demonstrate the efficacy of our approach in overcoming the limitations of supervised learning heavily reliant on large-scale labeled data.
3D Cross Pseudo Supervision : A semi-supervised nnU-Net architecture for abdominal organ segmentation
Sep 19, 2022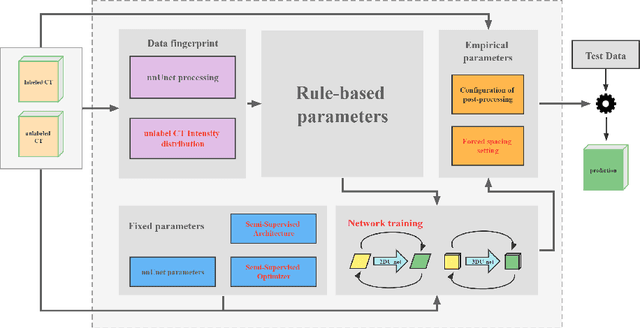
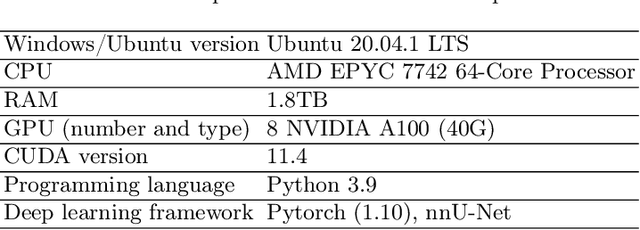
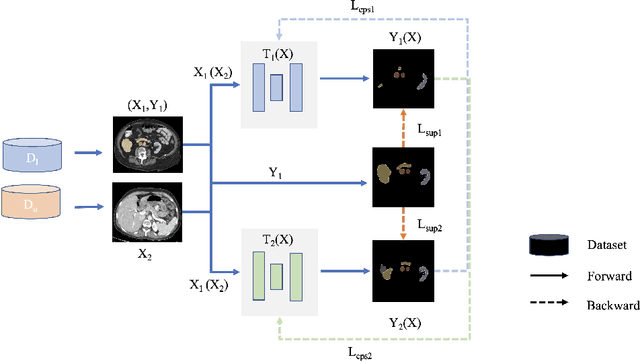
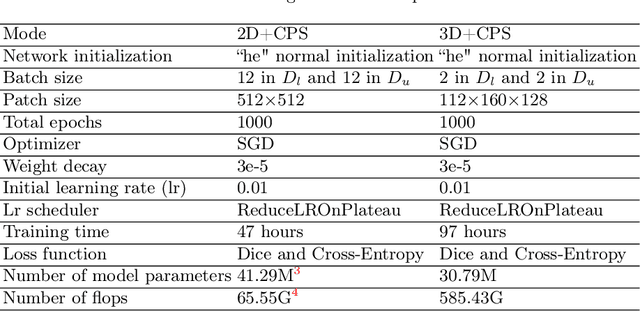
Abstract:Large curated datasets are necessary, but annotating medical images is a time-consuming, laborious, and expensive process. Therefore, recent supervised methods are focusing on utilizing a large amount of unlabeled data. However, to do so, is a challenging task. To address this problem, we propose a new 3D Cross Pseudo Supervision (3D-CPS) method, a semi-supervised network architecture based on nnU-Net with the Cross Pseudo Supervision method. We design a new nnU-Net based preprocessing method and adopt the forced spacing settings strategy in the inference stage to speed up the inference time. In addition, we set the semi-supervised loss weights to expand linearity with each epoch to prevent the model from low-quality pseudo-labels in the early training process. Our proposed method achieves an average dice similarity coefficient (DSC) of 0.881 and an average normalized surface distance (NSD) of 0.913 on the MICCAI FLARE2022 validation set (20 cases).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge