Haseeb Hassan
Label-efficient Multi-organ Segmentation Method with Diffusion Model
Feb 23, 2024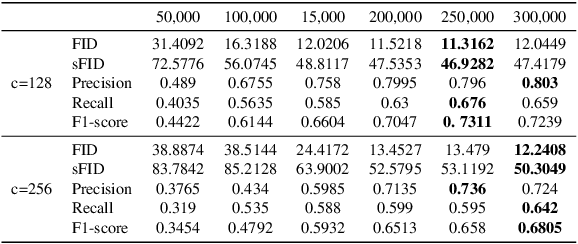
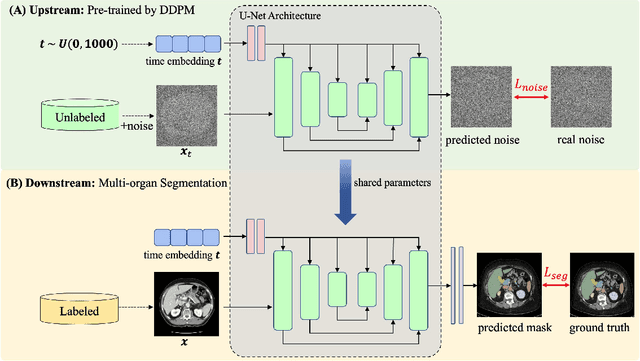

Abstract:Accurate segmentation of multiple organs in Computed Tomography (CT) images plays a vital role in computer-aided diagnosis systems. Various supervised-learning approaches have been proposed recently. However, these methods heavily depend on a large amount of high-quality labeled data, which is expensive to obtain in practice. In this study, we present a label-efficient learning approach using a pre-trained diffusion model for multi-organ segmentation tasks in CT images. First, a denoising diffusion model was trained using unlabeled CT data, generating additional two-dimensional (2D) CT images. Then the pre-trained denoising diffusion network was transferred to the downstream multi-organ segmentation task, effectively creating a semi-supervised learning model that requires only a small amount of labeled data. Furthermore, linear classification and fine-tuning decoder strategies were employed to enhance the network's segmentation performance. Our generative model at 256x256 resolution achieves impressive performance in terms of Fr\'echet inception distance, spatial Fr\'echet inception distance, and F1-score, with values of 11.32, 46.93, and 73.1\%, respectively. These results affirm the diffusion model's ability to generate diverse and realistic 2D CT images. Additionally, our method achieves competitive multi-organ segmentation performance compared to state-of-the-art methods on the FLARE 2022 dataset, particularly in limited labeled data scenarios. Remarkably, even with only 1\% and 10\% labeled data, our method achieves Dice similarity coefficients (DSCs) of 71.56\% and 78.51\% after fine-tuning, respectively. The method achieves a DSC score of 51.81\% using just four labeled CT scans. These results demonstrate the efficacy of our approach in overcoming the limitations of supervised learning heavily reliant on large-scale labeled data.
Quantitative perfusion maps using a novelty spatiotemporal convolutional neural network
Dec 08, 2023



Abstract:Dynamic susceptibility contrast magnetic resonance imaging (DSC-MRI) is widely used to evaluate acute ischemic stroke to distinguish salvageable tissue and infarct core. For this purpose, traditional methods employ deconvolution techniques, like singular value decomposition, which are known to be vulnerable to noise, potentially distorting the derived perfusion parameters. However, deep learning technology could leverage it, which can accurately estimate clinical perfusion parameters compared to traditional clinical approaches. Therefore, this study presents a perfusion parameters estimation network that considers spatial and temporal information, the Spatiotemporal Network (ST-Net), for the first time. The proposed network comprises a designed physical loss function to enhance model performance further. The results indicate that the network can accurately estimate perfusion parameters, including cerebral blood volume (CBV), cerebral blood flow (CBF), and time to maximum of the residual function (Tmax). The structural similarity index (SSIM) mean values for CBV, CBF, and Tmax parameters were 0.952, 0.943, and 0.863, respectively. The DICE score for the hypo-perfused region reached 0.859, demonstrating high consistency. The proposed model also maintains time efficiency, closely approaching the performance of commercial gold-standard software.
A Comparative Study of Pre-trained CNNs and GRU-Based Attention for Image Caption Generation
Oct 11, 2023



Abstract:Image captioning is a challenging task involving generating a textual description for an image using computer vision and natural language processing techniques. This paper proposes a deep neural framework for image caption generation using a GRU-based attention mechanism. Our approach employs multiple pre-trained convolutional neural networks as the encoder to extract features from the image and a GRU-based language model as the decoder to generate descriptive sentences. To improve performance, we integrate the Bahdanau attention model with the GRU decoder to enable learning to focus on specific image parts. We evaluate our approach using the MSCOCO and Flickr30k datasets and show that it achieves competitive scores compared to state-of-the-art methods. Our proposed framework can bridge the gap between computer vision and natural language and can be extended to specific domains.
Bayesian inference for data-efficient, explainable, and safe robotic motion planning: A review
Jul 16, 2023



Abstract:Bayesian inference has many advantages in robotic motion planning over four perspectives: The uncertainty quantification of the policy, safety (risk-aware) and optimum guarantees of robot motions, data-efficiency in training of reinforcement learning, and reducing the sim2real gap when the robot is applied to real-world tasks. However, the application of Bayesian inference in robotic motion planning is lagging behind the comprehensive theory of Bayesian inference. Further, there are no comprehensive reviews to summarize the progress of Bayesian inference to give researchers a systematic understanding in robotic motion planning. This paper first provides the probabilistic theories of Bayesian inference which are the preliminary of Bayesian inference for complex cases. Second, the Bayesian estimation is given to estimate the posterior of policies or unknown functions which are used to compute the policy. Third, the classical model-based Bayesian RL and model-free Bayesian RL algorithms for robotic motion planning are summarized, while these algorithms in complex cases are also analyzed. Fourth, the analysis of Bayesian inference in inverse RL is given to infer the reward functions in a data-efficient manner. Fifth, we systematically present the hybridization of Bayesian inference and RL which is a promising direction to improve the convergence of RL for better motion planning. Sixth, given the Bayesian inference, we present the interpretable and safe robotic motion plannings which are the hot research topic recently. Finally, all algorithms reviewed in this paper are summarized analytically as the knowledge graphs, and the future of Bayesian inference for robotic motion planning is also discussed, to pave the way for data-efficient, explainable, and safe robotic motion planning strategies for practical applications.
3D Cross Pseudo Supervision : A semi-supervised nnU-Net architecture for abdominal organ segmentation
Sep 19, 2022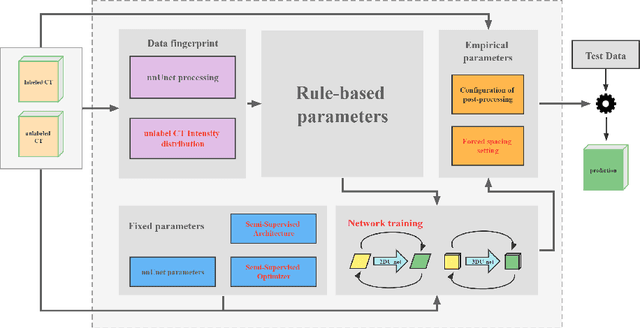
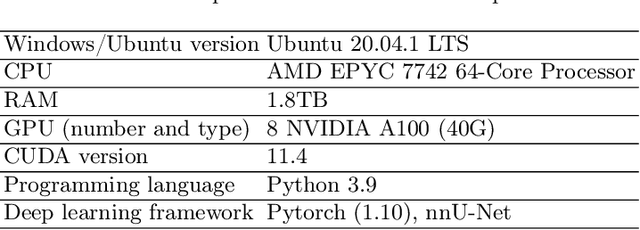
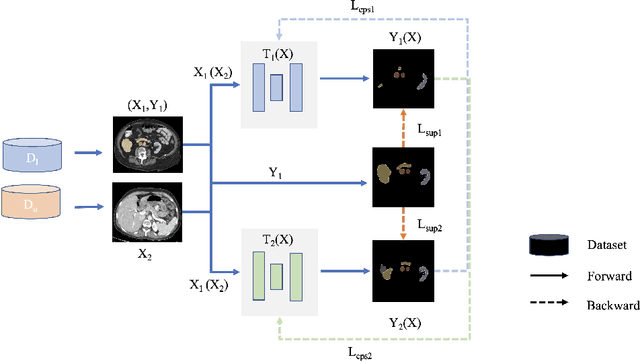
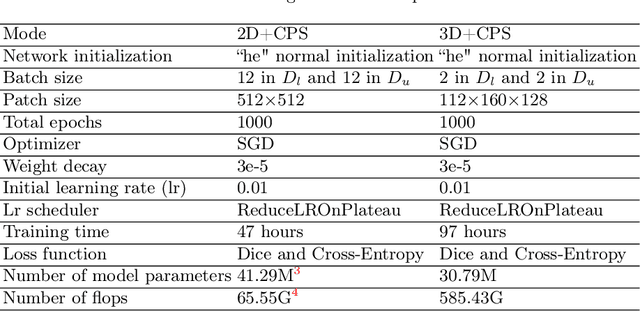
Abstract:Large curated datasets are necessary, but annotating medical images is a time-consuming, laborious, and expensive process. Therefore, recent supervised methods are focusing on utilizing a large amount of unlabeled data. However, to do so, is a challenging task. To address this problem, we propose a new 3D Cross Pseudo Supervision (3D-CPS) method, a semi-supervised network architecture based on nnU-Net with the Cross Pseudo Supervision method. We design a new nnU-Net based preprocessing method and adopt the forced spacing settings strategy in the inference stage to speed up the inference time. In addition, we set the semi-supervised loss weights to expand linearity with each epoch to prevent the model from low-quality pseudo-labels in the early training process. Our proposed method achieves an average dice similarity coefficient (DSC) of 0.881 and an average normalized surface distance (NSD) of 0.913 on the MICCAI FLARE2022 validation set (20 cases).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge