Xueqiang Zeng
Quantitative perfusion maps using a novelty spatiotemporal convolutional neural network
Dec 08, 2023



Abstract:Dynamic susceptibility contrast magnetic resonance imaging (DSC-MRI) is widely used to evaluate acute ischemic stroke to distinguish salvageable tissue and infarct core. For this purpose, traditional methods employ deconvolution techniques, like singular value decomposition, which are known to be vulnerable to noise, potentially distorting the derived perfusion parameters. However, deep learning technology could leverage it, which can accurately estimate clinical perfusion parameters compared to traditional clinical approaches. Therefore, this study presents a perfusion parameters estimation network that considers spatial and temporal information, the Spatiotemporal Network (ST-Net), for the first time. The proposed network comprises a designed physical loss function to enhance model performance further. The results indicate that the network can accurately estimate perfusion parameters, including cerebral blood volume (CBV), cerebral blood flow (CBF), and time to maximum of the residual function (Tmax). The structural similarity index (SSIM) mean values for CBV, CBF, and Tmax parameters were 0.952, 0.943, and 0.863, respectively. The DICE score for the hypo-perfused region reached 0.859, demonstrating high consistency. The proposed model also maintains time efficiency, closely approaching the performance of commercial gold-standard software.
Progressive Sampling-Based Bayesian Optimization for Efficient and Automatic Machine Learning Model Selection
Dec 06, 2018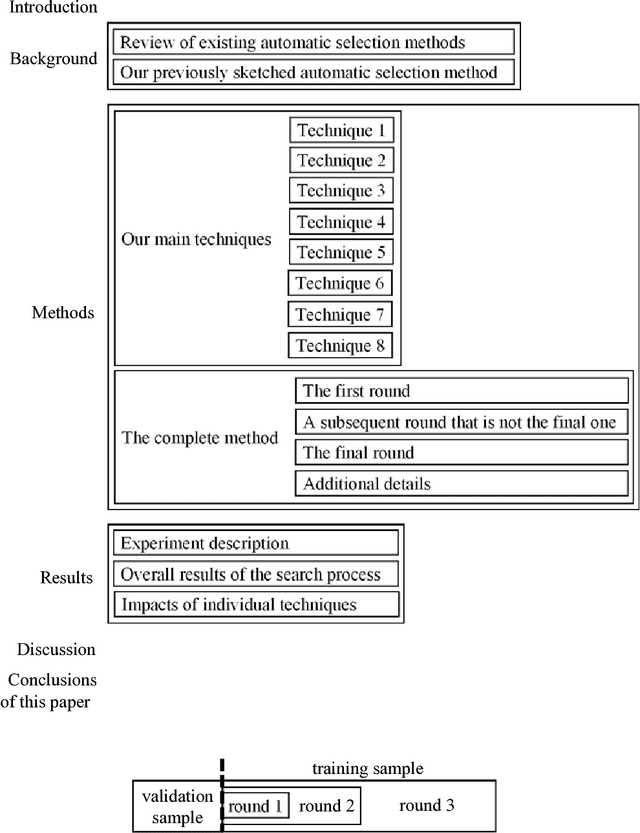
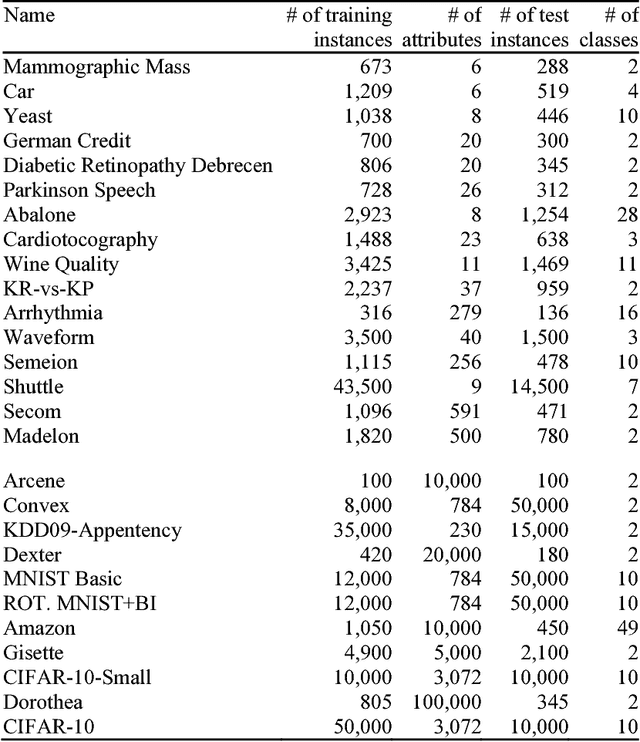
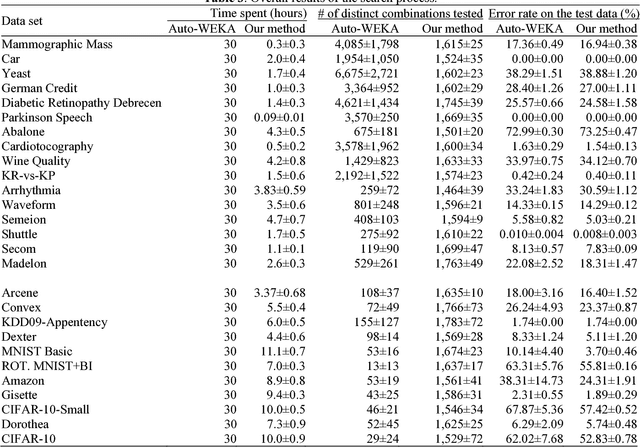
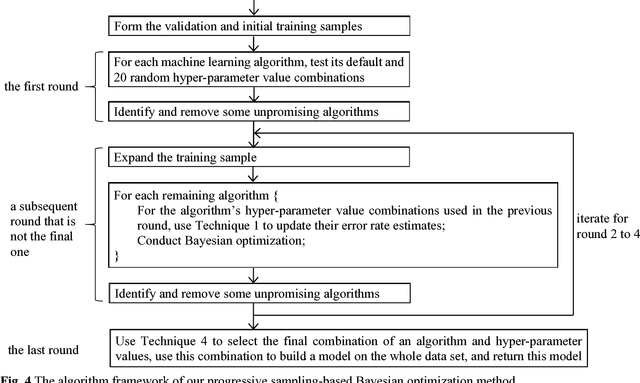
Abstract:Purpose: Machine learning is broadly used for clinical data analysis. Before training a model, a machine learning algorithm must be selected. Also, the values of one or more model parameters termed hyper-parameters must be set. Selecting algorithms and hyper-parameter values requires advanced machine learning knowledge and many labor-intensive manual iterations. To lower the bar to machine learning, miscellaneous automatic selection methods for algorithms and/or hyper-parameter values have been proposed. Existing automatic selection methods are inefficient on large data sets. This poses a challenge for using machine learning in the clinical big data era. Methods: To address the challenge, this paper presents progressive sampling-based Bayesian optimization, an efficient and automatic selection method for both algorithms and hyper-parameter values. Results: We report an implementation of the method. We show that compared to a state of the art automatic selection method, our method can significantly reduce search time, classification error rate, and standard deviation of error rate due to randomization. Conclusions: This is major progress towards enabling fast turnaround in identifying high-quality solutions required by many machine learning-based clinical data analysis tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge