Sen Xu
4D-CAAL: 4D Radar-Camera Calibration and Auto-Labeling for Autonomous Driving
Jan 29, 2026Abstract:4D radar has emerged as a critical sensor for autonomous driving, primarily due to its enhanced capabilities in elevation measurement and higher resolution compared to traditional 3D radar. Effective integration of 4D radar with cameras requires accurate extrinsic calibration, and the development of radar-based perception algorithms demands large-scale annotated datasets. However, existing calibration methods often employ separate targets optimized for either visual or radar modalities, complicating correspondence establishment. Furthermore, manually labeling sparse radar data is labor-intensive and unreliable. To address these challenges, we propose 4D-CAAL, a unified framework for 4D radar-camera calibration and auto-labeling. Our approach introduces a novel dual-purpose calibration target design, integrating a checkerboard pattern on the front surface for camera detection and a corner reflector at the center of the back surface for radar detection. We develop a robust correspondence matching algorithm that aligns the checkerboard center with the strongest radar reflection point, enabling accurate extrinsic calibration. Subsequently, we present an auto-labeling pipeline that leverages the calibrated sensor relationship to transfer annotations from camera-based segmentations to radar point clouds through geometric projection and multi-feature optimization. Extensive experiments demonstrate that our method achieves high calibration accuracy while significantly reducing manual annotation effort, thereby accelerating the development of robust multi-modal perception systems for autonomous driving.
Tiny Model, Big Logic: Diversity-Driven Optimization Elicits Large-Model Reasoning Ability in VibeThinker-1.5B
Nov 09, 2025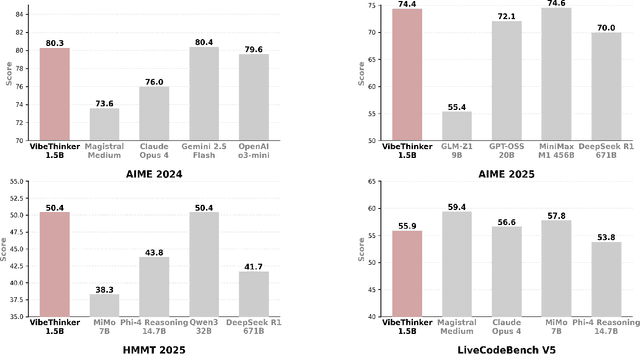
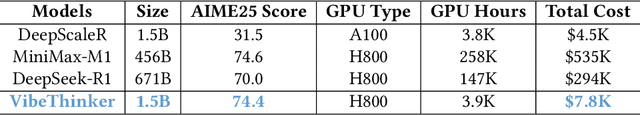
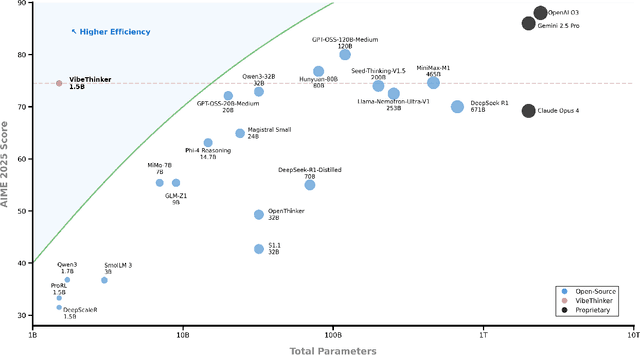
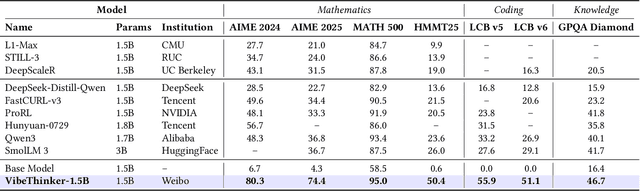
Abstract:Challenging the prevailing consensus that small models inherently lack robust reasoning, this report introduces VibeThinker-1.5B, a 1.5B-parameter dense model developed via our Spectrum-to-Signal Principle (SSP). This challenges the prevailing approach of scaling model parameters to enhance capabilities, as seen in models like DeepSeek R1 (671B) and Kimi k2 (>1T). The SSP framework first employs a Two-Stage Diversity-Exploring Distillation (SFT) to generate a broad spectrum of solutions, followed by MaxEnt-Guided Policy Optimization (RL) to amplify the correct signal. With a total training cost of only $7,800, VibeThinker-1.5B demonstrates superior reasoning capabilities compared to closed-source models like Magistral Medium and Claude Opus 4, and performs on par with open-source models like GPT OSS-20B Medium. Remarkably, it surpasses the 400x larger DeepSeek R1 on three math benchmarks: AIME24 (80.3 vs. 79.8), AIME25 (74.4 vs. 70.0), and HMMT25 (50.4 vs. 41.7). This is a substantial improvement over its base model (6.7, 4.3, and 0.6, respectively). On LiveCodeBench V6, it scores 51.1, outperforming Magistral Medium's 50.3 and its base model's 0.0. These findings demonstrate that small models can achieve reasoning capabilities comparable to large models, drastically reducing training and inference costs and thereby democratizing advanced AI research.
DALIP: Distribution Alignment-based Language-Image Pre-Training for Domain-Specific Data
Apr 02, 2025Abstract:Recently, Contrastive Language-Image Pre-training (CLIP) has shown promising performance in domain-specific data (e.g., biology), and has attracted increasing research attention. Existing works generally focus on collecting extensive domain-specific data and directly tuning the original CLIP models. Intuitively, such a paradigm takes no full consideration of the characteristics lying in domain-specific data (e.g., fine-grained nature of biological data) and so limits model capability, while mostly losing the original ability of CLIP in the general domain. In this paper, we propose a Distribution Alignment-based Language-Image Pre-Training (DALIP) method for biological data. Specifically, DALIP optimizes CLIP models by matching the similarity between feature distribution of image-text pairs instead of the original [cls] token, which can capture rich yet effective information inherent in image-text pairs as powerful representations, and so better cope with fine-grained nature of biological data. Particularly, our DALIP efficiently approximates feature distribution via its first- and second-order statistics, while presenting a Multi-head Brownian Distance Covariance (MBDC) module to acquire second-order statistics of token features efficiently. Furthermore, we collect a new dataset for plant domain (e.g., specific data in biological domain) comprising 10M plant data with 3M general-domain data (namely PlantMix-13M) according to data mixing laws. Extensive experiments show that DALIP clearly outperforms existing CLIP counterparts in biological domain, while well generalizing to remote sensing and medical imaging domains. Besides, our PlantMix-13M dataset further boosts performance of DALIP in plant domain, while preserving model ability in general domain.
Learning Invariant Inter-pixel Correlations for Superpixel Generation
Feb 28, 2024



Abstract:Deep superpixel algorithms have made remarkable strides by substituting hand-crafted features with learnable ones. Nevertheless, we observe that existing deep superpixel methods, serving as mid-level representation operations, remain sensitive to the statistical properties (e.g., color distribution, high-level semantics) embedded within the training dataset. Consequently, learnable features exhibit constrained discriminative capability, resulting in unsatisfactory pixel grouping performance, particularly in untrainable application scenarios. To address this issue, we propose the Content Disentangle Superpixel (CDS) algorithm to selectively separate the invariant inter-pixel correlations and statistical properties, i.e., style noise. Specifically, We first construct auxiliary modalities that are homologous to the original RGB image but have substantial stylistic variations. Then, driven by mutual information, we propose the local-grid correlation alignment across modalities to reduce the distribution discrepancy of adaptively selected features and learn invariant inter-pixel correlations. Afterwards, we perform global-style mutual information minimization to enforce the separation of invariant content and train data styles. The experimental results on four benchmark datasets demonstrate the superiority of our approach to existing state-of-the-art methods, regarding boundary adherence, generalization, and efficiency. Code and pre-trained model are available at https://github.com/rookiie/CDSpixel.
Ins-ATP: Deep Estimation of ATP for Organoid Based on High Throughput Microscopic Images
Mar 15, 2023



Abstract:Adenosine triphosphate (ATP) is a high-energy phosphate compound and the most direct energy source in organisms. ATP is an essential biomarker for evaluating cell viability in biology. Researchers often use ATP bioluminescence to measure the ATP of organoid after drug to evaluate the drug efficacy. However, ATP bioluminescence has some limitations, leading to unreliable drug screening results. Performing ATP bioluminescence causes cell lysis of organoids, so it is impossible to observe organoids' long-term viability changes after medication continually. To overcome the disadvantages of ATP bioluminescence, we propose Ins-ATP, a non-invasive strategy, the first organoid ATP estimation model based on the high-throughput microscopic image. Ins-ATP directly estimates the ATP of organoids from high-throughput microscopic images, so that it does not influence the drug reactions of organoids. Therefore, the ATP change of organoids can be observed for a long time to obtain more stable results. Experimental results show that the ATP estimation by Ins-ATP is in good agreement with those determined by ATP bioluminescence. Specifically, the predictions of Ins-ATP are consistent with the results measured by ATP bioluminescence in the efficacy evaluation experiments of different drugs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge