Jin Keun Seo
Neural Representation-Based Method for Metal-induced Artifact Reduction in Dental CBCT Imaging
Jul 27, 2023Abstract:This study introduces a novel reconstruction method for dental cone-beam computed tomography (CBCT), focusing on effectively reducing metal-induced artifacts commonly encountered in the presence of prevalent metallic implants. Despite significant progress in metal artifact reduction techniques, challenges persist owing to the intricate physical interactions between polychromatic X-ray beams and metal objects, which are further compounded by the additional effects associated with metal-tooth interactions and factors specific to the dental CBCT data environment. To overcome these limitations, we propose an implicit neural network that generates two distinct and informative tomographic images. One image represents the monochromatic attenuation distribution at a specific energy level, whereas the other captures the nonlinear beam-hardening factor resulting from the polychromatic nature of X-ray beams. In contrast to existing CT reconstruction techniques, the proposed method relies exclusively on the Beer--Lambert law, effectively preventing the generation of metal-induced artifacts during the backprojection process commonly implemented in conventional methods. Extensive experimental evaluations demonstrate that the proposed method effectively reduces metal artifacts while providing high-quality image reconstructions, thus emphasizing the significance of the second image in capturing the nonlinear beam-hardening factor.
Automatic 3D Registration of Dental CBCT and Face Scan Data using 2D Projection images
May 17, 2023



Abstract:This paper presents a fully automatic registration method of dental cone-beam computed tomography (CBCT) and face scan data. It can be used for a digital platform of 3D jaw-teeth-face models in a variety of applications, including 3D digital treatment planning and orthognathic surgery. Difficulties in accurately merging facial scans and CBCT images are due to the different image acquisition methods and limited area of correspondence between the two facial surfaces. In addition, it is difficult to use machine learning techniques because they use face-related 3D medical data with radiation exposure, which are difficult to obtain for training. The proposed method addresses these problems by reusing an existing machine-learning-based 2D landmark detection algorithm in an open-source library and developing a novel mathematical algorithm that identifies paired 3D landmarks from knowledge of the corresponding 2D landmarks. A main contribution of this study is that the proposed method does not require annotated training data of facial landmarks because it uses a pre-trained facial landmark detection algorithm that is known to be robust and generalized to various 2D face image models. Note that this reduces a 3D landmark detection problem to a 2D problem of identifying the corresponding landmarks on two 2D projection images generated from two different projection angles. Here, the 3D landmarks for registration were selected from the sub-surfaces with the least geometric change under the CBCT and face scan environments. For the final fine-tuning of the registration, the Iterative Closest Point method was applied, which utilizes geometrical information around the 3D landmarks. The experimental results show that the proposed method achieved an averaged surface distance error of 0.74 mm for three pairs of CBCT and face scan datasets.
Nonlinear ill-posed problem in low-dose dental cone-beam computed tomography
Mar 03, 2023



Abstract:This paper describes the mathematical structure of the ill-posed nonlinear inverse problem of low-dose dental cone-beam computed tomography (CBCT) and explains the advantages of a deep learning-based approach to the reconstruction of computed tomography images over conventional regularization methods. This paper explains the underlying reasons why dental CBCT is more ill-posed than standard computed tomography. Despite this severe ill-posedness, the demand for dental CBCT systems is rapidly growing because of their cost competitiveness and low radiation dose. We then describe the limitations of existing methods in the accurate restoration of the morphological structures of teeth using dental CBCT data severely damaged by metal implants. We further discuss the usefulness of panoramic images generated from CBCT data for accurate tooth segmentation. We also discuss the possibility of utilizing radiation-free intra-oral scan data as prior information in CBCT image reconstruction to compensate for the damage to data caused by metal implants.
Metal Artifact Reduction with Intra-Oral Scan Data for 3D Low Dose Maxillofacial CBCT Modeling
Feb 08, 2022



Abstract:Low-dose dental cone beam computed tomography (CBCT) has been increasingly used for maxillofacial modeling. However, the presence of metallic inserts, such as implants, crowns, and dental filling, causes severe streaking and shading artifacts in a CBCT image and loss of the morphological structures of the teeth, which consequently prevents accurate segmentation of bones. A two-stage metal artifact reduction method is proposed for accurate 3D low-dose maxillofacial CBCT modeling, where a key idea is to utilize explicit tooth shape prior information from intra-oral scan data whose acquisition does not require any extra radiation exposure. In the first stage, an image-to-image deep learning network is employed to mitigate metal-related artifacts. To improve the learning ability, the proposed network is designed to take advantage of the intra-oral scan data as side-inputs and perform multi-task learning of auxiliary tooth segmentation. In the second stage, a 3D maxillofacial model is constructed by segmenting the bones from the dental CBCT image corrected in the first stage. For accurate bone segmentation, weighted thresholding is applied, wherein the weighting region is determined depending on the geometry of the intra-oral scan data. Because acquiring a paired training dataset of metal-artifact-free and metal artifact-affected dental CBCT images is challenging in clinical practice, an automatic method of generating a realistic dataset according to the CBCT physics model is introduced. Numerical simulations and clinical experiments show the feasibility of the proposed method, which takes advantage of tooth surface information from intra-oral scan data in 3D low dose maxillofacial CBCT modeling.
Fully automatic integration of dental CBCT images and full-arch intraoral impressions with stitching error correction via individual tooth segmentation and identification
Dec 03, 2021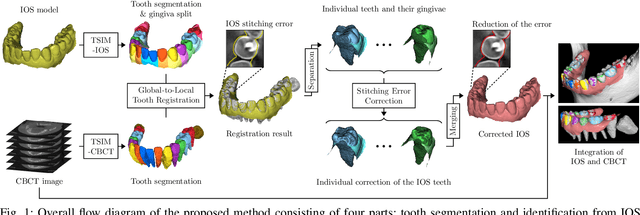
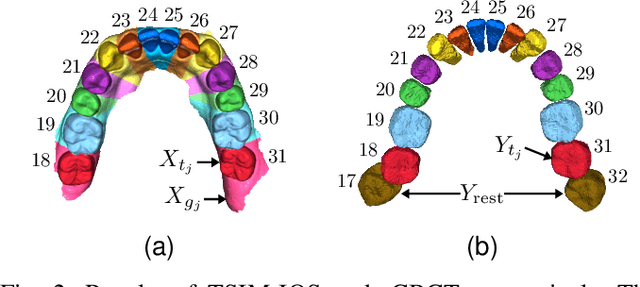
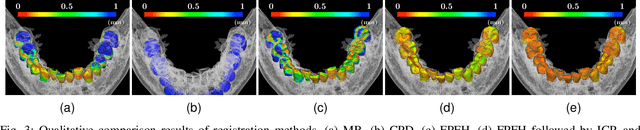
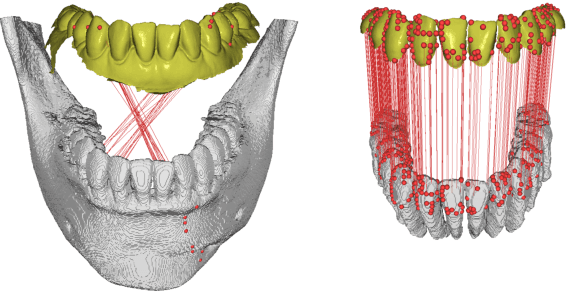
Abstract:We present a fully automated method of integrating intraoral scan (IOS) and dental cone-beam computerized tomography (CBCT) images into one image by complementing each image's weaknesses. Dental CBCT alone may not be able to delineate precise details of the tooth surface due to limited image resolution and various CBCT artifacts, including metal-induced artifacts. IOS is very accurate for the scanning of narrow areas, but it produces cumulative stitching errors during full-arch scanning. The proposed method is intended not only to compensate the low-quality of CBCT-derived tooth surfaces with IOS, but also to correct the cumulative stitching errors of IOS across the entire dental arch. Moreover, the integration provide both gingival structure of IOS and tooth roots of CBCT in one image. The proposed fully automated method consists of four parts; (i) individual tooth segmentation and identification module for IOS data (TSIM-IOS); (ii) individual tooth segmentation and identification module for CBCT data (TSIM-CBCT); (iii) global-to-local tooth registration between IOS and CBCT; and (iv) stitching error correction of full-arch IOS. The experimental results show that the proposed method achieved landmark and surface distance errors of 0.11mm and 0.30mm, respectively.
A fully automated method for 3D individual tooth identification and segmentation in dental CBCT
Feb 11, 2021Abstract:Accurate and automatic segmentation of three-dimensional (3D) individual teeth from cone-beam computerized tomography (CBCT) images is a challenging problem because of the difficulty in separating an individual tooth from adjacent teeth and its surrounding alveolar bone. Thus, this paper proposes a fully automated method of identifying and segmenting 3D individual teeth from dental CBCT images. The proposed method addresses the aforementioned difficulty by developing a deep learning-based hierarchical multi-step model. First, it automatically generates upper and lower jaws panoramic images to overcome the computational complexity caused by high-dimensional data and the curse of dimensionality associated with limited training dataset. The obtained 2D panoramic images are then used to identify 2D individual teeth and capture loose- and tight- regions of interest (ROIs) of 3D individual teeth. Finally, accurate 3D individual tooth segmentation is achieved using both loose and tight ROIs. Experimental results showed that the proposed method achieved an F1-score of 93.35% for tooth identification and a Dice similarity coefficient of 94.79% for individual 3D tooth segmentation. The results demonstrate that the proposed method provides an effective clinical and practical framework for digital dentistry.
Automated 3D cephalometric landmark identification using computerized tomography
Dec 16, 2020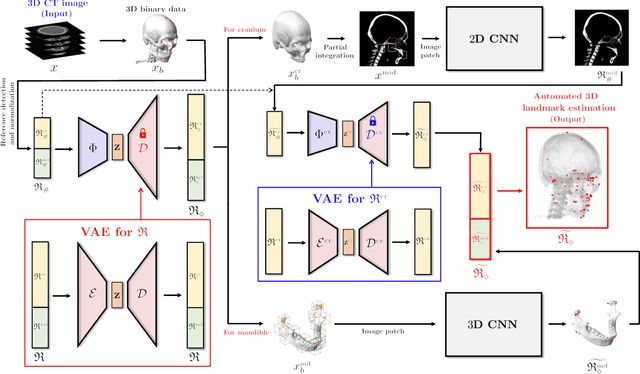
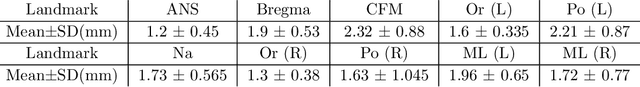
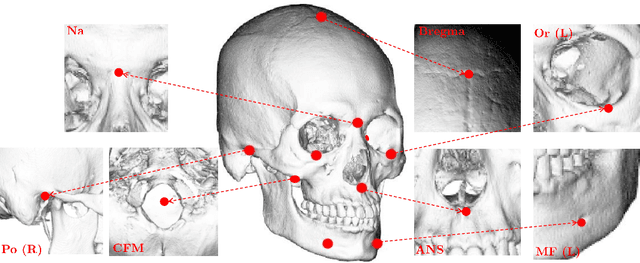
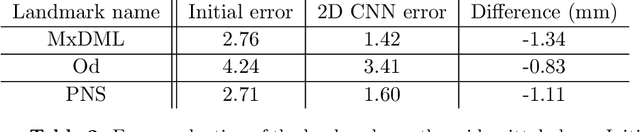
Abstract:Identification of 3D cephalometric landmarks that serve as proxy to the shape of human skull is the fundamental step in cephalometric analysis. Since manual landmarking from 3D computed tomography (CT) images is a cumbersome task even for the trained experts, automatic 3D landmark detection system is in a great need. Recently, automatic landmarking of 2D cephalograms using deep learning (DL) has achieved great success, but 3D landmarking for more than 80 landmarks has not yet reached a satisfactory level, because of the factors hindering machine learning such as the high dimensionality of the input data and limited amount of training data due to ethical restrictions on the use of medical data. This paper presents a semi-supervised DL method for 3D landmarking that takes advantage of anonymized landmark dataset with paired CT data being removed. The proposed method first detects a small number of easy-to-find reference landmarks, then uses them to provide a rough estimation of the entire landmarks by utilizing the low dimensional representation learned by variational autoencoder (VAE). Anonymized landmark dataset is used for training the VAE. Finally, coarse-to-fine detection is applied to the small bounding box provided by rough estimation, using separate strategies suitable for mandible and cranium. For mandibular landmarks, patch-based 3D CNN is applied to the segmented image of the mandible (separated from the maxilla), in order to capture 3D morphological features of mandible associated with the landmarks. We detect 6 landmarks around the condyle all at once, instead of one by one, because they are closely related to each other. For cranial landmarks, we again use VAE-based latent representation for more accurate annotation. In our experiment, the proposed method achieved an averaged 3D point-to-point error of 2.91 mm for 90 landmarks only with 15 paired training data.
Deep Learning-Based Solvability of Underdetermined Inverse Problems in Medical Imaging
Jan 07, 2020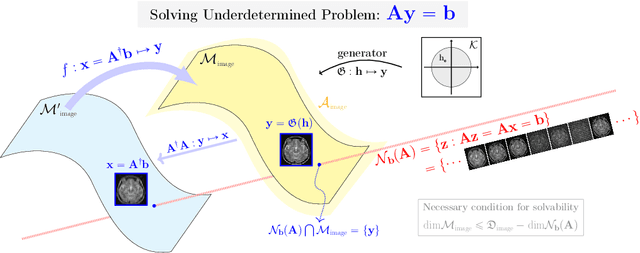
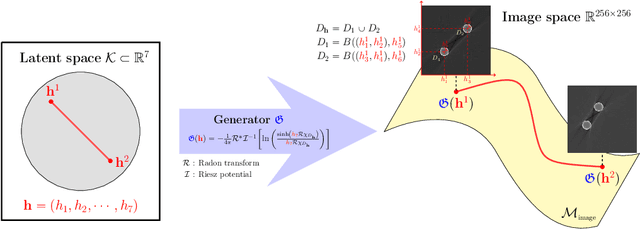
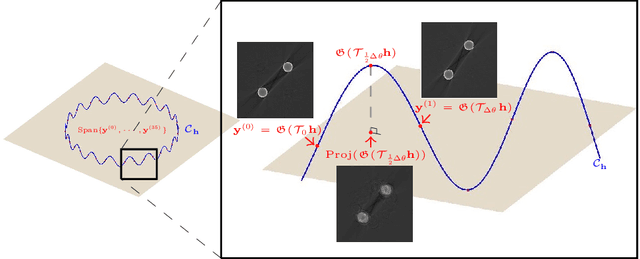
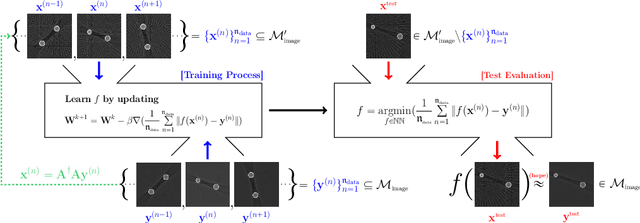
Abstract:Recently, with the significant developments in deep learning techniques, solving underdetermined inverse problems has become one of the major concerns in the medical imaging domain. Typical examples include undersampled magnetic resonance imaging, interior tomography, and sparse-view computed tomography, where deep learning techniques have achieved excellent performances. Although deep learning methods appear to overcome the limitations of existing mathematical methods when handling various underdetermined problems, there is a lack of rigorous mathematical foundations that would allow us to elucidate the reasons for the remarkable performance of deep learning methods. This study focuses on learning the causal relationship regarding the structure of the training data suitable for deep learning, to solve highly underdetermined inverse problems. We observe that a majority of the problems of solving underdetermined linear systems in medical imaging are highly non-linear. Furthermore, we analyze if a desired reconstruction map can be learnable from the training data and underdetermined system.
Framelet Pooling Aided Deep Learning Network : The Method to Process High Dimensional Medical Data
Jul 25, 2019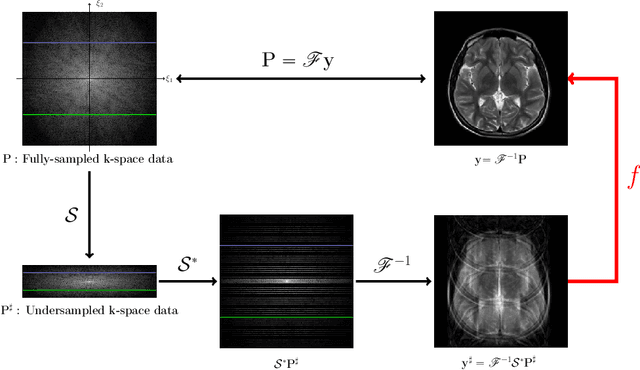
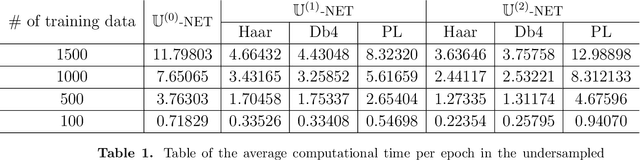
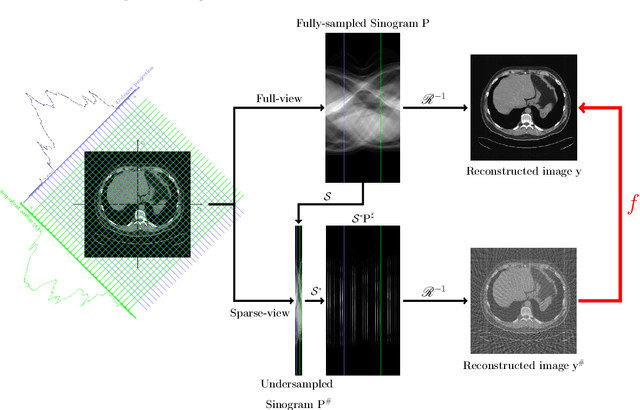
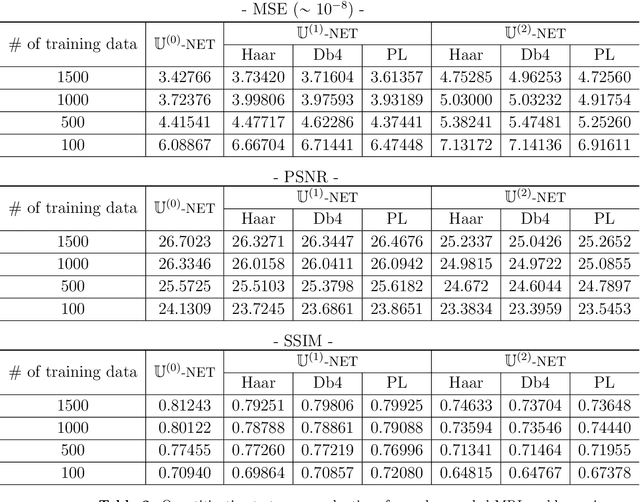
Abstract:Machine learning-based analysis of medical images often faces several hurdles, such as the lack of training data, the curse of dimensionality problem, and the generalization issues. One of the main difficulties is that there exists computational cost problem in dealing with input data of large size matrices which represent medical images. The purpose of this paper is to introduce a framelet-pooling aided deep learning method for mitigating computational bundle, caused by large dimensionality. By transforming high dimensional data into low dimensional components by filter banks with preserving detailed information, the proposed method aims to reduce the complexity of the neural network and computational costs significantly during the learning process. Various experiments show that our method is comparable to the standard unreduced learning method, while reducing computational burdens by decomposing large-sized learning tasks into several small-scale learning tasks.
Unpaired image denoising using a generative adversarial network in X-ray CT
Mar 04, 2019
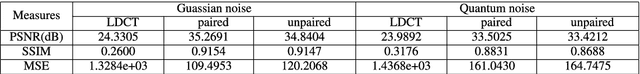
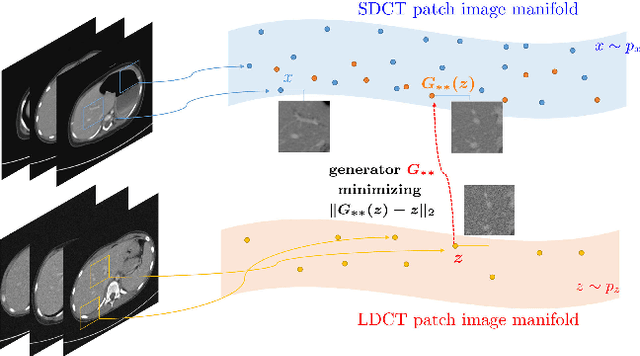
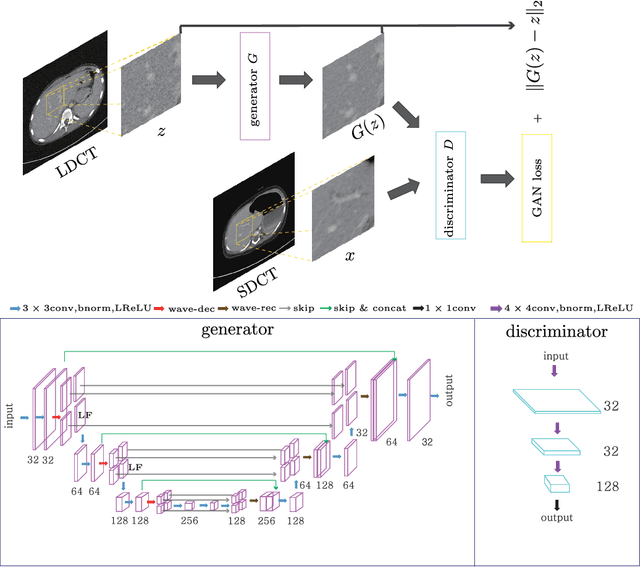
Abstract:This paper proposes a deep learning-based denoising method for noisy low-dose computerized tomography (CT) images in the absence of paired training data. The proposed method uses a fidelity-embedded generative adversarial network (GAN) to learn a denoising function from unpaired training data of low-dose CT (LDCT) and standard-dose CT (SDCT) images, where the denoising function is the optimal generator in the GAN framework. Given an optimal discriminator in the GAN, the generator is optimized by minimizing a weighted sum of two losses: the Kullback-Leibler divergence between an SDCT data distribution and a generated distribution, and the $\ell_2$ loss between the LDCT image and the corresponding generated images (or denoised image). The experimental results show that the proposed deep-learning method with unpaired datasets performs comparably to a method using paired datasets. Clinical experiment was also performed to show the validity of the proposed method for non-Gaussian noise arising in the low-dose X-ray CT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge