Seong Hyeon Baek
Automated 3D cephalometric landmark identification using computerized tomography
Dec 16, 2020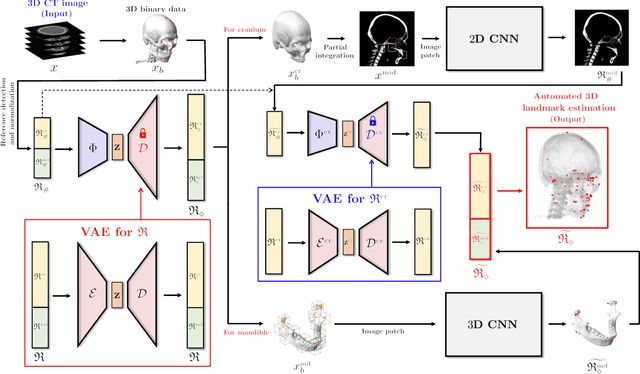
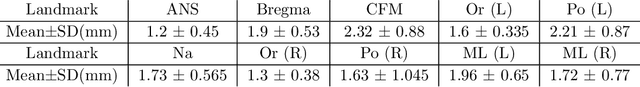
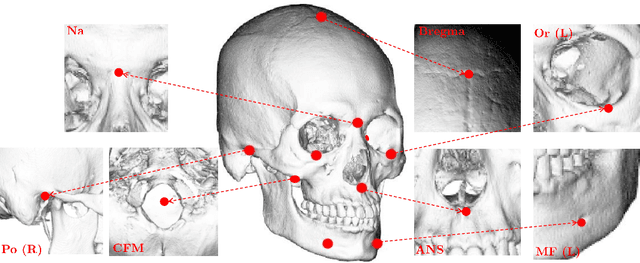
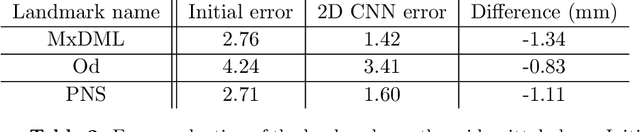
Abstract:Identification of 3D cephalometric landmarks that serve as proxy to the shape of human skull is the fundamental step in cephalometric analysis. Since manual landmarking from 3D computed tomography (CT) images is a cumbersome task even for the trained experts, automatic 3D landmark detection system is in a great need. Recently, automatic landmarking of 2D cephalograms using deep learning (DL) has achieved great success, but 3D landmarking for more than 80 landmarks has not yet reached a satisfactory level, because of the factors hindering machine learning such as the high dimensionality of the input data and limited amount of training data due to ethical restrictions on the use of medical data. This paper presents a semi-supervised DL method for 3D landmarking that takes advantage of anonymized landmark dataset with paired CT data being removed. The proposed method first detects a small number of easy-to-find reference landmarks, then uses them to provide a rough estimation of the entire landmarks by utilizing the low dimensional representation learned by variational autoencoder (VAE). Anonymized landmark dataset is used for training the VAE. Finally, coarse-to-fine detection is applied to the small bounding box provided by rough estimation, using separate strategies suitable for mandible and cranium. For mandibular landmarks, patch-based 3D CNN is applied to the segmented image of the mandible (separated from the maxilla), in order to capture 3D morphological features of mandible associated with the landmarks. We detect 6 landmarks around the condyle all at once, instead of one by one, because they are closely related to each other. For cranial landmarks, we again use VAE-based latent representation for more accurate annotation. In our experiment, the proposed method achieved an averaged 3D point-to-point error of 2.91 mm for 90 landmarks only with 15 paired training data.
Deep Learning-Based Solvability of Underdetermined Inverse Problems in Medical Imaging
Jan 07, 2020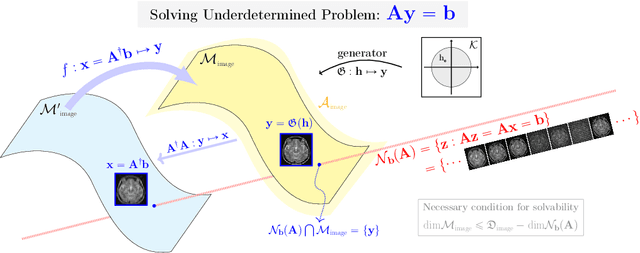
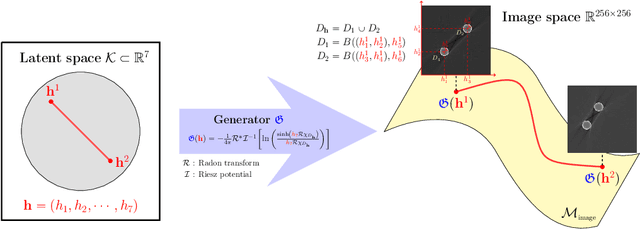
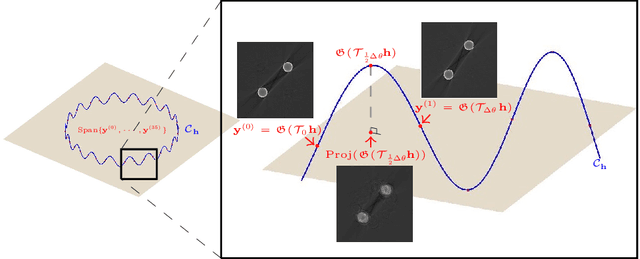
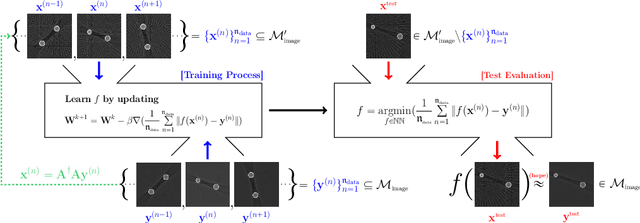
Abstract:Recently, with the significant developments in deep learning techniques, solving underdetermined inverse problems has become one of the major concerns in the medical imaging domain. Typical examples include undersampled magnetic resonance imaging, interior tomography, and sparse-view computed tomography, where deep learning techniques have achieved excellent performances. Although deep learning methods appear to overcome the limitations of existing mathematical methods when handling various underdetermined problems, there is a lack of rigorous mathematical foundations that would allow us to elucidate the reasons for the remarkable performance of deep learning methods. This study focuses on learning the causal relationship regarding the structure of the training data suitable for deep learning, to solve highly underdetermined inverse problems. We observe that a majority of the problems of solving underdetermined linear systems in medical imaging are highly non-linear. Furthermore, we analyze if a desired reconstruction map can be learnable from the training data and underdetermined system.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge