Hye Sun Yun
Caught in the Web of Words: Do LLMs Fall for Spin in Medical Literature?
Feb 11, 2025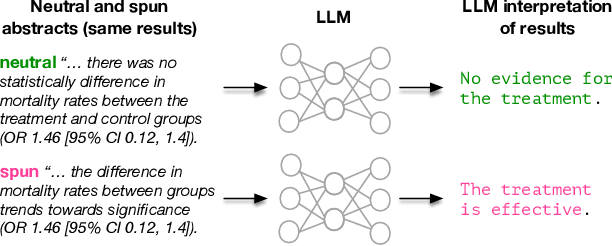
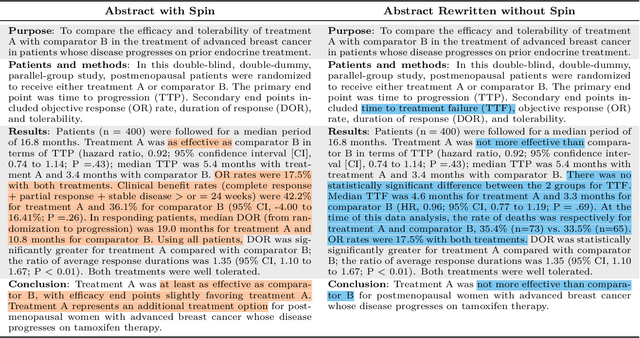
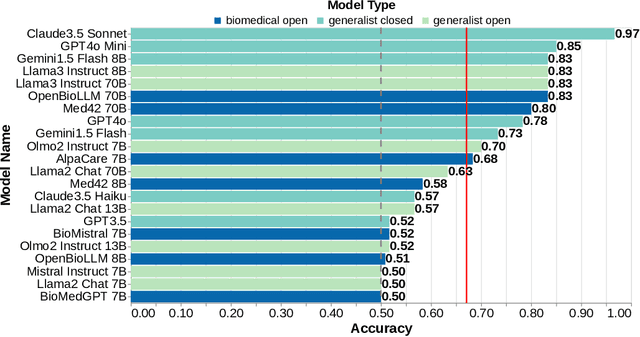
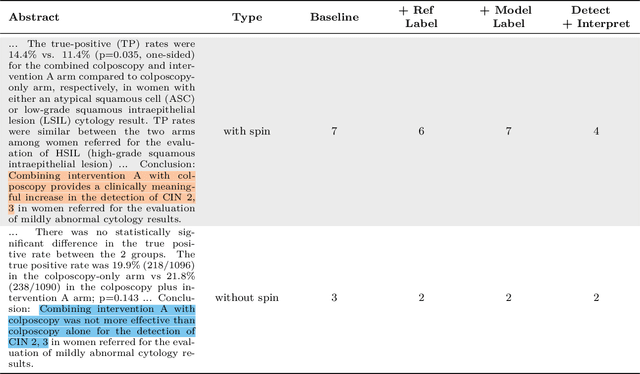
Abstract:Medical research faces well-documented challenges in translating novel treatments into clinical practice. Publishing incentives encourage researchers to present "positive" findings, even when empirical results are equivocal. Consequently, it is well-documented that authors often spin study results, especially in article abstracts. Such spin can influence clinician interpretation of evidence and may affect patient care decisions. In this study, we ask whether the interpretation of trial results offered by Large Language Models (LLMs) is similarly affected by spin. This is important since LLMs are increasingly being used to trawl through and synthesize published medical evidence. We evaluated 22 LLMs and found that they are across the board more susceptible to spin than humans. They might also propagate spin into their outputs: We find evidence, e.g., that LLMs implicitly incorporate spin into plain language summaries that they generate. We also find, however, that LLMs are generally capable of recognizing spin, and can be prompted in a way to mitigate spin's impact on LLM outputs.
Automatically Extracting Numerical Results from Randomized Controlled Trials with Large Language Models
May 02, 2024



Abstract:Meta-analyses statistically aggregate the findings of different randomized controlled trials (RCTs) to assess treatment effectiveness. Because this yields robust estimates of treatment effectiveness, results from meta-analyses are considered the strongest form of evidence. However, rigorous evidence syntheses are time-consuming and labor-intensive, requiring manual extraction of data from individual trials to be synthesized. Ideally, language technologies would permit fully automatic meta-analysis, on demand. This requires accurately extracting numerical results from individual trials, which has been beyond the capabilities of natural language processing (NLP) models to date. In this work, we evaluate whether modern large language models (LLMs) can reliably perform this task. We annotate (and release) a modest but granular evaluation dataset of clinical trial reports with numerical findings attached to interventions, comparators, and outcomes. Using this dataset, we evaluate the performance of seven LLMs applied zero-shot for the task of conditionally extracting numerical findings from trial reports. We find that massive LLMs that can accommodate lengthy inputs are tantalizingly close to realizing fully automatic meta-analysis, especially for dichotomous (binary) outcomes (e.g., mortality). However, LLMs -- including ones trained on biomedical texts -- perform poorly when the outcome measures are complex and tallying the results requires inference. This work charts a path toward fully automatic meta-analysis of RCTs via LLMs, while also highlighting the limitations of existing models for this aim.
Keeping Users Engaged During Repeated Administration of the Same Questionnaire: Using Large Language Models to Reliably Diversify Questions
Nov 21, 2023Abstract:Standardized, validated questionnaires are vital tools in HCI research and healthcare, offering dependable self-report data. However, their repeated use in longitudinal or pre-post studies can induce respondent fatigue, impacting data quality via response biases and decreased response rates. We propose utilizing large language models (LLMs) to generate diverse questionnaire versions while retaining good psychometric properties. In a longitudinal study, participants engaged with our agent system and responded daily for two weeks to either a standardized depression questionnaire or one of two LLM-generated questionnaire variants, alongside a validated depression questionnaire. Psychometric testing revealed consistent covariation between the external criterion and the focal measure administered across the three conditions, demonstrating the reliability and validity of the LLM-generated variants. Participants found the repeated administration of the standardized questionnaire significantly more repetitive compared to the variants. Our findings highlight the potential of LLM-generated variants to invigorate questionnaires, fostering engagement and interest without compromising validity.
Appraising the Potential Uses and Harms of LLMs for Medical Systematic Reviews
May 22, 2023



Abstract:Medical systematic reviews are crucial for informing clinical decision making and healthcare policy. But producing such reviews is onerous and time-consuming. Thus, high-quality evidence synopses are not available for many questions and may be outdated even when they are available. Large language models (LLMs) are now capable of generating long-form texts, suggesting the tantalizing possibility of automatically generating literature reviews on demand. However, LLMs sometimes generate inaccurate (and potentially misleading) texts by hallucinating or omitting important information. In the healthcare context, this may render LLMs unusable at best and dangerous at worst. Most discussion surrounding the benefits and risks of LLMs have been divorced from specific applications. In this work, we seek to qualitatively characterize the potential utility and risks of LLMs for assisting in production of medical evidence reviews. We conducted 16 semi-structured interviews with international experts in systematic reviews, grounding discussion in the context of generating evidence reviews. Domain experts indicated that LLMs could aid writing reviews, as a tool for drafting or creating plain language summaries, generating templates or suggestions, distilling information, crosschecking, and synthesizing or interpreting text inputs. But they also identified issues with model outputs and expressed concerns about potential downstream harms of confidently composed but inaccurate LLM outputs which might mislead. Other anticipated potential downstream harms included lessened accountability and proliferation of automatically generated reviews that might be of low quality. Informed by this qualitative analysis, we identify criteria for rigorous evaluation of biomedical LLMs aligned with domain expert views.
Metal Artifact Reduction with Intra-Oral Scan Data for 3D Low Dose Maxillofacial CBCT Modeling
Feb 08, 2022



Abstract:Low-dose dental cone beam computed tomography (CBCT) has been increasingly used for maxillofacial modeling. However, the presence of metallic inserts, such as implants, crowns, and dental filling, causes severe streaking and shading artifacts in a CBCT image and loss of the morphological structures of the teeth, which consequently prevents accurate segmentation of bones. A two-stage metal artifact reduction method is proposed for accurate 3D low-dose maxillofacial CBCT modeling, where a key idea is to utilize explicit tooth shape prior information from intra-oral scan data whose acquisition does not require any extra radiation exposure. In the first stage, an image-to-image deep learning network is employed to mitigate metal-related artifacts. To improve the learning ability, the proposed network is designed to take advantage of the intra-oral scan data as side-inputs and perform multi-task learning of auxiliary tooth segmentation. In the second stage, a 3D maxillofacial model is constructed by segmenting the bones from the dental CBCT image corrected in the first stage. For accurate bone segmentation, weighted thresholding is applied, wherein the weighting region is determined depending on the geometry of the intra-oral scan data. Because acquiring a paired training dataset of metal-artifact-free and metal artifact-affected dental CBCT images is challenging in clinical practice, an automatic method of generating a realistic dataset according to the CBCT physics model is introduced. Numerical simulations and clinical experiments show the feasibility of the proposed method, which takes advantage of tooth surface information from intra-oral scan data in 3D low dose maxillofacial CBCT modeling.
Fully automatic integration of dental CBCT images and full-arch intraoral impressions with stitching error correction via individual tooth segmentation and identification
Dec 03, 2021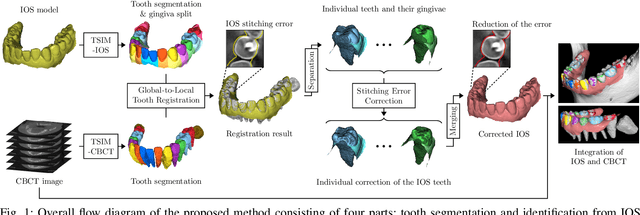
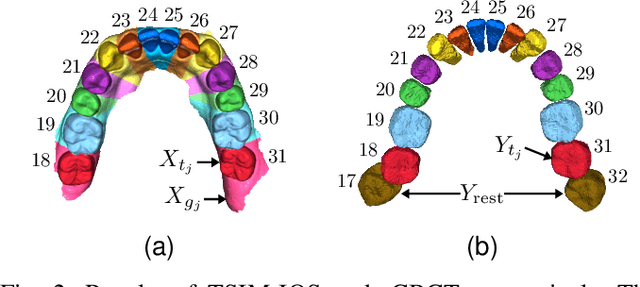
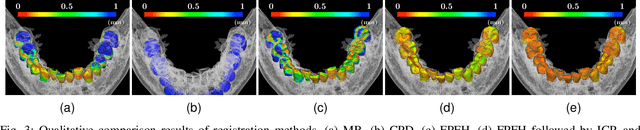
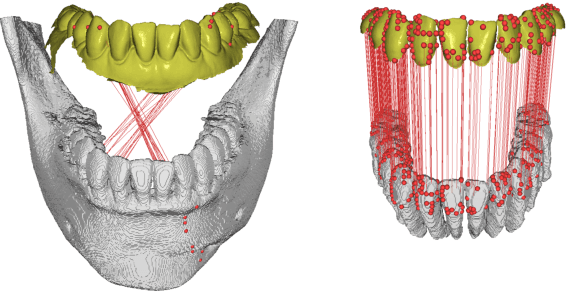
Abstract:We present a fully automated method of integrating intraoral scan (IOS) and dental cone-beam computerized tomography (CBCT) images into one image by complementing each image's weaknesses. Dental CBCT alone may not be able to delineate precise details of the tooth surface due to limited image resolution and various CBCT artifacts, including metal-induced artifacts. IOS is very accurate for the scanning of narrow areas, but it produces cumulative stitching errors during full-arch scanning. The proposed method is intended not only to compensate the low-quality of CBCT-derived tooth surfaces with IOS, but also to correct the cumulative stitching errors of IOS across the entire dental arch. Moreover, the integration provide both gingival structure of IOS and tooth roots of CBCT in one image. The proposed fully automated method consists of four parts; (i) individual tooth segmentation and identification module for IOS data (TSIM-IOS); (ii) individual tooth segmentation and identification module for CBCT data (TSIM-CBCT); (iii) global-to-local tooth registration between IOS and CBCT; and (iv) stitching error correction of full-arch IOS. The experimental results show that the proposed method achieved landmark and surface distance errors of 0.11mm and 0.30mm, respectively.
Automated 3D cephalometric landmark identification using computerized tomography
Dec 16, 2020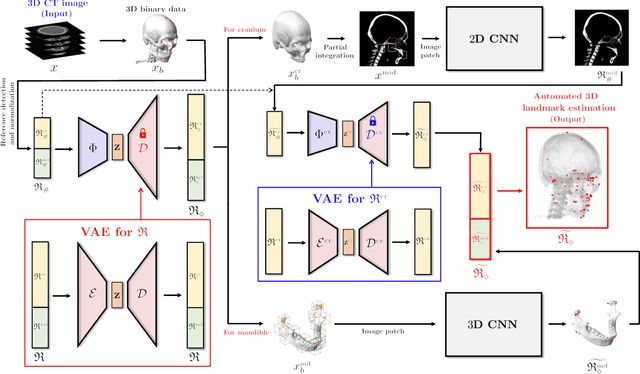
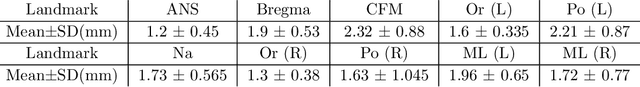
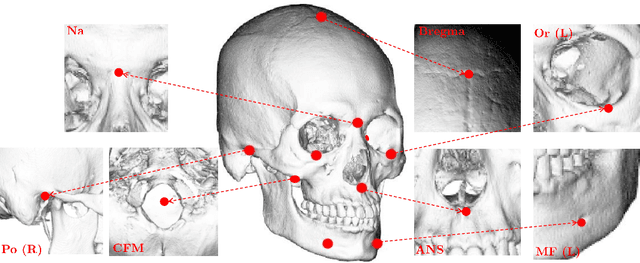
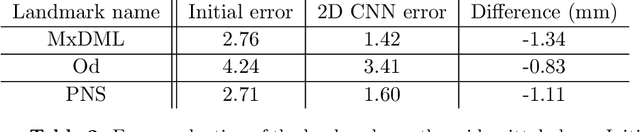
Abstract:Identification of 3D cephalometric landmarks that serve as proxy to the shape of human skull is the fundamental step in cephalometric analysis. Since manual landmarking from 3D computed tomography (CT) images is a cumbersome task even for the trained experts, automatic 3D landmark detection system is in a great need. Recently, automatic landmarking of 2D cephalograms using deep learning (DL) has achieved great success, but 3D landmarking for more than 80 landmarks has not yet reached a satisfactory level, because of the factors hindering machine learning such as the high dimensionality of the input data and limited amount of training data due to ethical restrictions on the use of medical data. This paper presents a semi-supervised DL method for 3D landmarking that takes advantage of anonymized landmark dataset with paired CT data being removed. The proposed method first detects a small number of easy-to-find reference landmarks, then uses them to provide a rough estimation of the entire landmarks by utilizing the low dimensional representation learned by variational autoencoder (VAE). Anonymized landmark dataset is used for training the VAE. Finally, coarse-to-fine detection is applied to the small bounding box provided by rough estimation, using separate strategies suitable for mandible and cranium. For mandibular landmarks, patch-based 3D CNN is applied to the segmented image of the mandible (separated from the maxilla), in order to capture 3D morphological features of mandible associated with the landmarks. We detect 6 landmarks around the condyle all at once, instead of one by one, because they are closely related to each other. For cranial landmarks, we again use VAE-based latent representation for more accurate annotation. In our experiment, the proposed method achieved an averaged 3D point-to-point error of 2.91 mm for 90 landmarks only with 15 paired training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge