Tae Jun Jang
NeRF Solves Undersampled MRI Reconstruction
Feb 20, 2024



Abstract:This article presents a novel undersampled magnetic resonance imaging (MRI) technique that leverages the concept of Neural Radiance Field (NeRF). With radial undersampling, the corresponding imaging problem can be reformulated into an image modeling task from sparse-view rendered data; therefore, a high dimensional MR image is obtainable from undersampled $k$-space data by taking advantage of implicit neural representation. A multi-layer perceptron, which is designed to output an image intensity from a spatial coordinate, learns the MR physics-driven rendering relation between given measurement data and desired image. Effective undersampling strategies for high-quality neural representation are investigated. The proposed method serves two benefits: (i) The learning is based fully on single undersampled $k$-space data, not a bunch of measured data and target image sets. It can be used potentially for diagnostic MR imaging, such as fetal MRI, where data acquisition is relatively rare or limited against diversity of clinical images while undersampled reconstruction is highly demanded. (ii) A reconstructed MR image is a scan-specific representation highly adaptive to the given $k$-space measurement. Numerous experiments validate the feasibility and capability of the proposed approach.
Machine Learning-based Signal Quality Assessment for Cardiac Volume Monitoring in Electrical Impedance Tomography
Jan 04, 2023Abstract:Owing to recent advances in thoracic electrical impedance tomography, a patient's hemodynamic function can be noninvasively and continuously estimated in real-time by surveilling a cardiac volume signal associated with stroke volume and cardiac output. In clinical applications, however, a cardiac volume signal is often of low quality, mainly because of the patient's deliberate movements or inevitable motions during clinical interventions. This study aims to develop a signal quality indexing method that assesses the influence of motion artifacts on transient cardiac volume signals. The assessment is performed on each cardiac cycle to take advantage of the periodicity and regularity in cardiac volume changes. Time intervals are identified using the synchronized electrocardiography system. We apply divergent machine-learning methods, which can be sorted into discriminative-model and manifold-learning approaches. The use of machine-learning could be suitable for our real-time monitoring application that requires fast inference and automation as well as high accuracy. In the clinical environment, the proposed method can be utilized to provide immediate warnings so that clinicians can minimize confusion regarding patients' conditions, reduce clinical resource utilization, and improve the confidence level of the monitoring system. Numerous experiments using actual EIT data validate the capability of cardiac volume signals degraded by motion artifacts to be accurately and automatically assessed in real-time by machine learning. The best model achieved an accuracy of 0.95, positive and negative predictive values of 0.96 and 0.86, sensitivity of 0.98, specificity of 0.77, and AUC of 0.96.
Metal Artifact Reduction with Intra-Oral Scan Data for 3D Low Dose Maxillofacial CBCT Modeling
Feb 08, 2022



Abstract:Low-dose dental cone beam computed tomography (CBCT) has been increasingly used for maxillofacial modeling. However, the presence of metallic inserts, such as implants, crowns, and dental filling, causes severe streaking and shading artifacts in a CBCT image and loss of the morphological structures of the teeth, which consequently prevents accurate segmentation of bones. A two-stage metal artifact reduction method is proposed for accurate 3D low-dose maxillofacial CBCT modeling, where a key idea is to utilize explicit tooth shape prior information from intra-oral scan data whose acquisition does not require any extra radiation exposure. In the first stage, an image-to-image deep learning network is employed to mitigate metal-related artifacts. To improve the learning ability, the proposed network is designed to take advantage of the intra-oral scan data as side-inputs and perform multi-task learning of auxiliary tooth segmentation. In the second stage, a 3D maxillofacial model is constructed by segmenting the bones from the dental CBCT image corrected in the first stage. For accurate bone segmentation, weighted thresholding is applied, wherein the weighting region is determined depending on the geometry of the intra-oral scan data. Because acquiring a paired training dataset of metal-artifact-free and metal artifact-affected dental CBCT images is challenging in clinical practice, an automatic method of generating a realistic dataset according to the CBCT physics model is introduced. Numerical simulations and clinical experiments show the feasibility of the proposed method, which takes advantage of tooth surface information from intra-oral scan data in 3D low dose maxillofacial CBCT modeling.
Fully automatic integration of dental CBCT images and full-arch intraoral impressions with stitching error correction via individual tooth segmentation and identification
Dec 03, 2021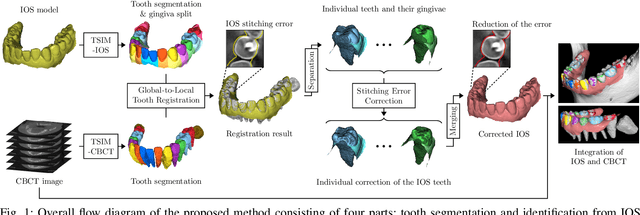
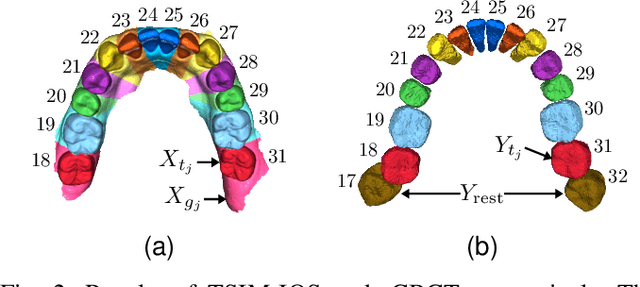
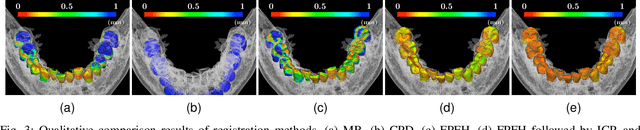
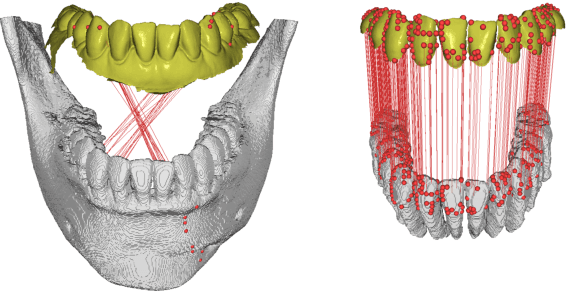
Abstract:We present a fully automated method of integrating intraoral scan (IOS) and dental cone-beam computerized tomography (CBCT) images into one image by complementing each image's weaknesses. Dental CBCT alone may not be able to delineate precise details of the tooth surface due to limited image resolution and various CBCT artifacts, including metal-induced artifacts. IOS is very accurate for the scanning of narrow areas, but it produces cumulative stitching errors during full-arch scanning. The proposed method is intended not only to compensate the low-quality of CBCT-derived tooth surfaces with IOS, but also to correct the cumulative stitching errors of IOS across the entire dental arch. Moreover, the integration provide both gingival structure of IOS and tooth roots of CBCT in one image. The proposed fully automated method consists of four parts; (i) individual tooth segmentation and identification module for IOS data (TSIM-IOS); (ii) individual tooth segmentation and identification module for CBCT data (TSIM-CBCT); (iii) global-to-local tooth registration between IOS and CBCT; and (iv) stitching error correction of full-arch IOS. The experimental results show that the proposed method achieved landmark and surface distance errors of 0.11mm and 0.30mm, respectively.
A fully automated method for 3D individual tooth identification and segmentation in dental CBCT
Feb 11, 2021Abstract:Accurate and automatic segmentation of three-dimensional (3D) individual teeth from cone-beam computerized tomography (CBCT) images is a challenging problem because of the difficulty in separating an individual tooth from adjacent teeth and its surrounding alveolar bone. Thus, this paper proposes a fully automated method of identifying and segmenting 3D individual teeth from dental CBCT images. The proposed method addresses the aforementioned difficulty by developing a deep learning-based hierarchical multi-step model. First, it automatically generates upper and lower jaws panoramic images to overcome the computational complexity caused by high-dimensional data and the curse of dimensionality associated with limited training dataset. The obtained 2D panoramic images are then used to identify 2D individual teeth and capture loose- and tight- regions of interest (ROIs) of 3D individual teeth. Finally, accurate 3D individual tooth segmentation is achieved using both loose and tight ROIs. Experimental results showed that the proposed method achieved an F1-score of 93.35% for tooth identification and a Dice similarity coefficient of 94.79% for individual 3D tooth segmentation. The results demonstrate that the proposed method provides an effective clinical and practical framework for digital dentistry.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge