Jijun Tang
Dual-Channel Reliable Breast Ultrasound Image Classification Based on Explainable Attribution and Uncertainty Quantification
Jan 08, 2024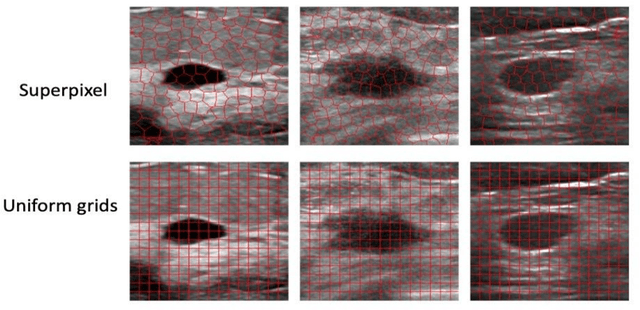
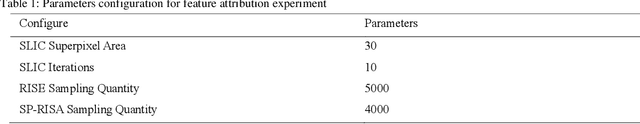
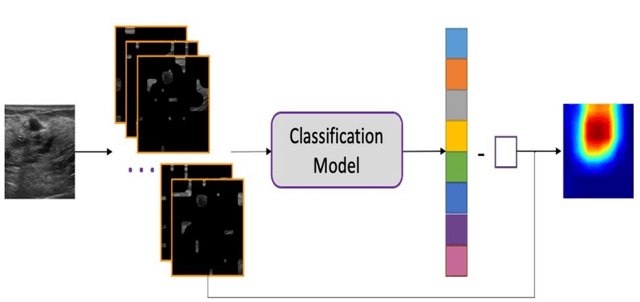

Abstract:This paper focuses on the classification task of breast ultrasound images and researches on the reliability measurement of classification results. We proposed a dual-channel evaluation framework based on the proposed inference reliability and predictive reliability scores. For the inference reliability evaluation, human-aligned and doctor-agreed inference rationales based on the improved feature attribution algorithm SP-RISA are gracefully applied. Uncertainty quantification is used to evaluate the predictive reliability via the Test Time Enhancement. The effectiveness of this reliability evaluation framework has been verified on our breast ultrasound clinical dataset YBUS, and its robustness is verified on the public dataset BUSI. The expected calibration errors on both datasets are significantly lower than traditional evaluation methods, which proves the effectiveness of our proposed reliability measurement.
EAA-Net: Rethinking the Autoencoder Architecture with Intra-class Features for Medical Image Segmentation
Aug 19, 2022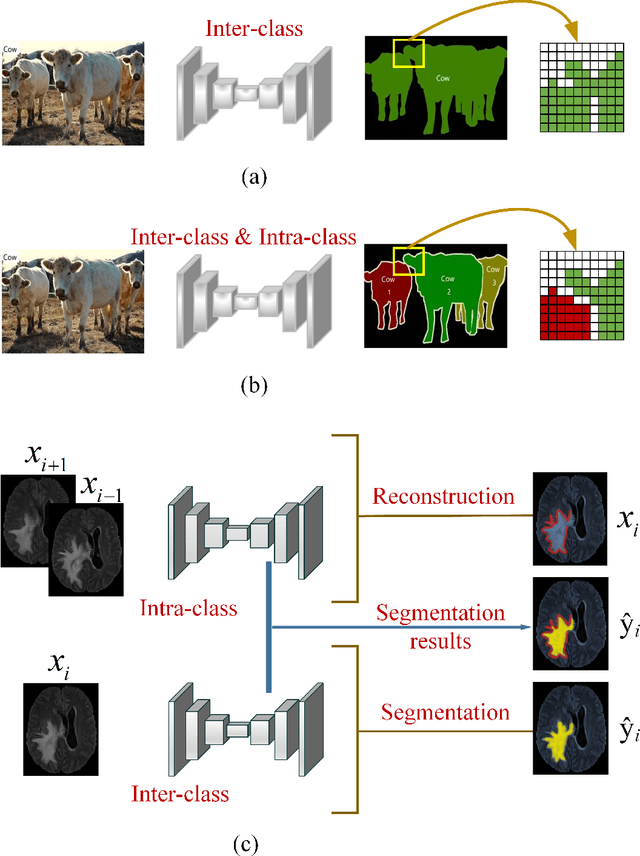
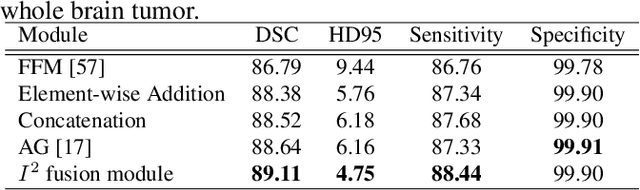
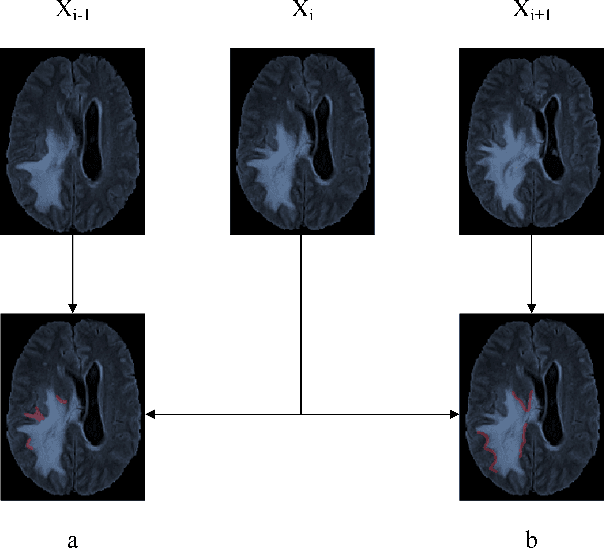
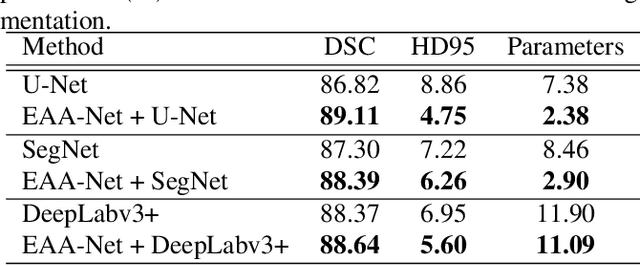
Abstract:Automatic image segmentation technology is critical to the visual analysis. The autoencoder architecture has satisfying performance in various image segmentation tasks. However, autoencoders based on convolutional neural networks (CNN) seem to encounter a bottleneck in improving the accuracy of semantic segmentation. Increasing the inter-class distance between foreground and background is an inherent characteristic of the segmentation network. However, segmentation networks pay too much attention to the main visual difference between foreground and background, and ignores the detailed edge information, which leads to a reduction in the accuracy of edge segmentation. In this paper, we propose a light-weight end-to-end segmentation framework based on multi-task learning, termed Edge Attention autoencoder Network (EAA-Net), to improve edge segmentation ability. Our approach not only utilizes the segmentation network to obtain inter-class features, but also applies the reconstruction network to extract intra-class features among the foregrounds. We further design a intra-class and inter-class features fusion module -- I2 fusion module. The I2 fusion module is used to merge intra-class and inter-class features, and use a soft attention mechanism to remove invalid background information. Experimental results show that our method performs well in medical image segmentation tasks. EAA-Net is easy to implement and has small calculation cost.
TranSiam: Fusing Multimodal Visual Features Using Transformer for Medical Image Segmentation
Apr 26, 2022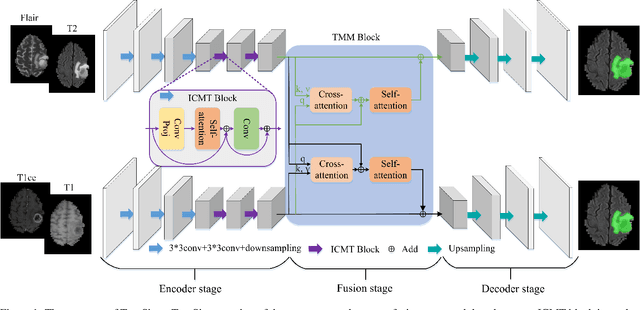
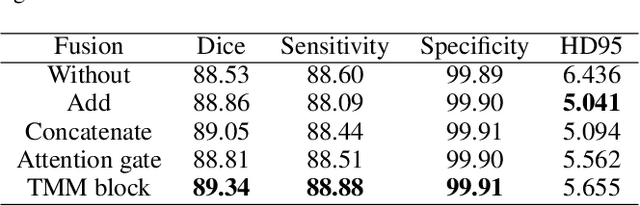
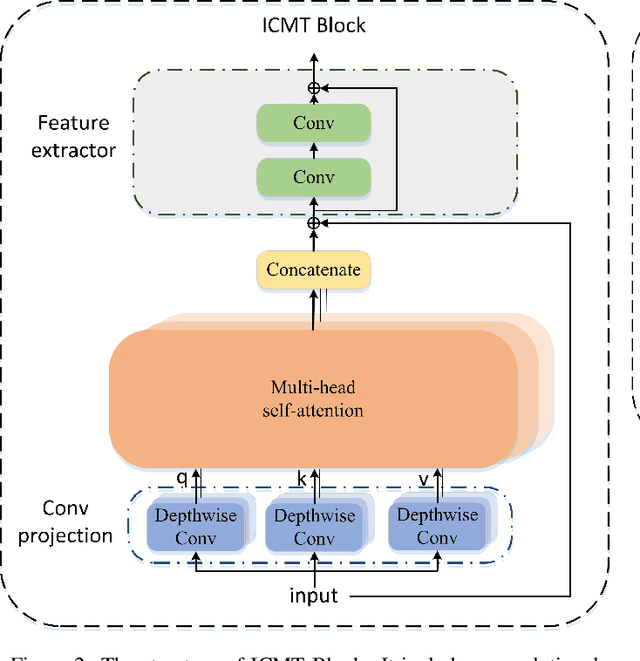
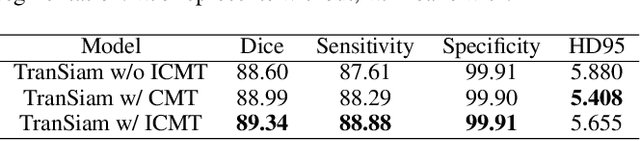
Abstract:Automatic segmentation of medical images based on multi-modality is an important topic for disease diagnosis. Although the convolutional neural network (CNN) has been proven to have excellent performance in image segmentation tasks, it is difficult to obtain global information. The lack of global information will seriously affect the accuracy of the segmentation results of the lesion area. In addition, there are visual representation differences between multimodal data of the same patient. These differences will affect the results of the automatic segmentation methods. To solve these problems, we propose a segmentation method suitable for multimodal medical images that can capture global information, named TranSiam. TranSiam is a 2D dual path network that extracts features of different modalities. In each path, we utilize convolution to extract detailed information in low level stage, and design a ICMT block to extract global information in high level stage. ICMT block embeds convolution in the transformer, which can extract global information while retaining spatial and detailed information. Furthermore, we design a novel fusion mechanism based on cross attention and selfattention, called TMM block, which can effectively fuse features between different modalities. On the BraTS 2019 and BraTS 2020 multimodal datasets, we have a significant improvement in accuracy over other popular methods.
Pretata: predicting TATA binding proteins with novel features and dimensionality reduction strategy
Mar 07, 2017
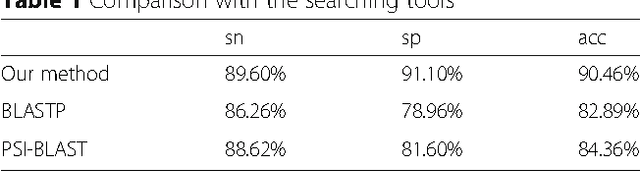
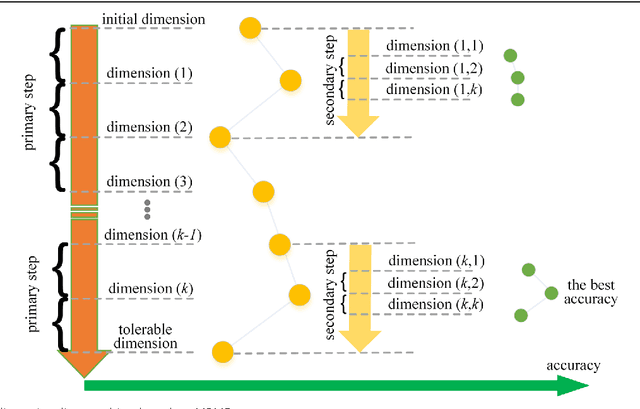
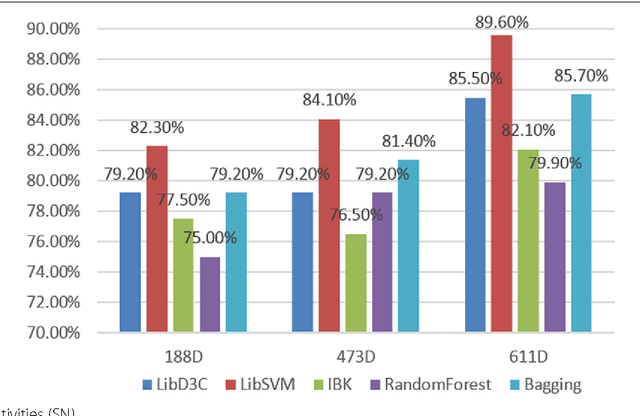
Abstract:Background: It is necessary and essential to discovery protein function from the novel primary sequences. Wet lab experimental procedures are not only time-consuming, but also costly, so predicting protein structure and function reliably based only on amino acid sequence has significant value. TATA-binding protein (TBP) is a kind of DNA binding protein, which plays a key role in the transcription regulation. Our study proposed an automatic approach for identifying TATA-binding proteins efficiently, accurately, and conveniently. This method would guide for the special protein identification with computational intelligence strategies. Results: Firstly, we proposed novel fingerprint features for TBP based on pseudo amino acid composition, physicochemical properties, and secondary structure. Secondly, hierarchical features dimensionality reduction strategies were employed to improve the performance furthermore. Currently, Pretata achieves 92.92% TATA- binding protein prediction accuracy, which is better than all other existing methods. Conclusions: The experiments demonstrate that our method could greatly improve the prediction accuracy and speed, thus allowing large-scale NGS data prediction to be practical. A web server is developed to facilitate the other researchers, which can be accessed at http://server.malab.cn/preTata/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge