Ioannis Tsamardinos
University of Crete
A Comparative Analysis of Influence Signals for Data Debugging
Jun 13, 2025Abstract:Improving the quality of training samples is crucial for improving the reliability and performance of ML models. In this paper, we conduct a comparative evaluation of influence-based signals for debugging training data. These signals can potentially identify both mislabeled and anomalous samples from a potentially noisy training set as we build the models and hence alleviate the need for dedicated glitch detectors. Although several influence-based signals (e.g., Self-Influence, Average Absolute Influence, Marginal Influence, GD-class) have been recently proposed in the literature, there are no experimental studies for assessing their power in detecting different glitch types (e.g., mislabeled and anomalous samples) under a common influence estimator (e.g., TraceIn) for different data modalities (image and tabular), and deep learning models (trained from scratch or foundation). Through extensive experiments, we show that signals like Self-Influence effectively detect mislabeled samples, but none of the existing signals can detect anomalies. Existing signals do not take into account the training dynamics, i.e., how the samples' influence on the model changes during training, while some signals fall into influence cancellation effects, i.e., influence score is zero due to unsigned scores accumulation, resulting in misleading influence attribution.
Confidence Interval Estimation of Predictive Performance in the Context of AutoML
Jun 12, 2024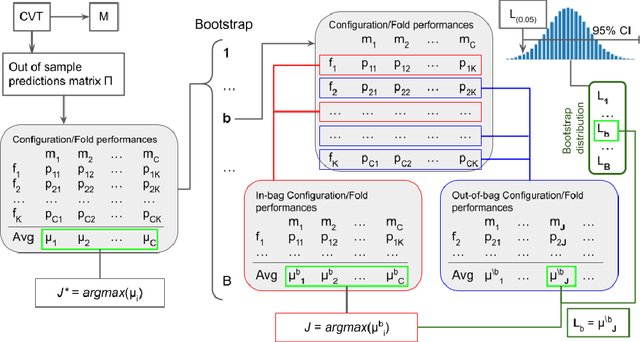
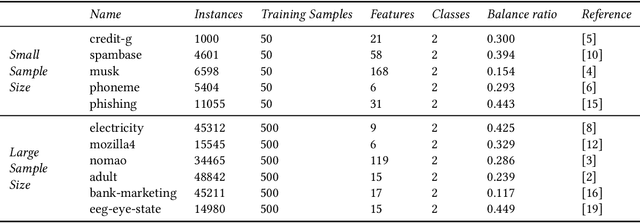
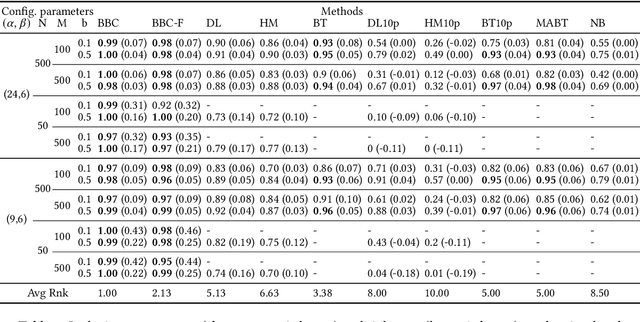
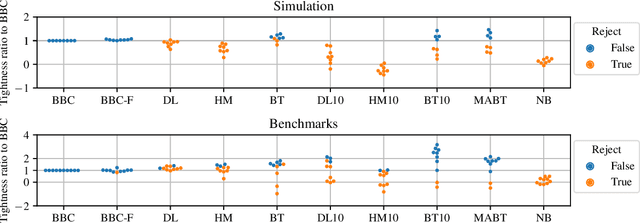
Abstract:Any supervised machine learning analysis is required to provide an estimate of the out-of-sample predictive performance. However, it is imperative to also provide a quantification of the uncertainty of this performance in the form of a confidence or credible interval (CI) and not just a point estimate. In an AutoML setting, estimating the CI is challenging due to the ``winner's curse", i.e., the bias of estimation due to cross-validating several machine learning pipelines and selecting the winning one. In this work, we perform a comparative evaluation of 9 state-of-the-art methods and variants in CI estimation in an AutoML setting on a corpus of real and simulated datasets. The methods are compared in terms of inclusion percentage (does a 95\% CI include the true performance at least 95\% of the time), CI tightness (tighter CIs are preferable as being more informative), and execution time. The evaluation is the first one that covers most, if not all, such methods and extends previous work to imbalanced and small-sample tasks. In addition, we present a variant, called BBC-F, of an existing method (the Bootstrap Bias Correction, or BBC) that maintains the statistical properties of the BBC but is more computationally efficient. The results support that BBC-F and BBC dominate the other methods in all metrics measured.
Towards Automated Causal Discovery: a case study on 5G telecommunication data
Feb 22, 2024Abstract:We introduce the concept of Automated Causal Discovery (AutoCD), defined as any system that aims to fully automate the application of causal discovery and causal reasoning methods. AutoCD's goal is to deliver all causal information that an expert human analyst would and answer a user's causal queries. We describe the architecture of such a platform, and illustrate its performance on synthetic data sets. As a case study, we apply it on temporal telecommunication data. The system is general and can be applied to a plethora of causal discovery problems.
A Meta-Level Learning Algorithm for Sequential Hyper-Parameter Space Reduction in AutoML
Dec 11, 2023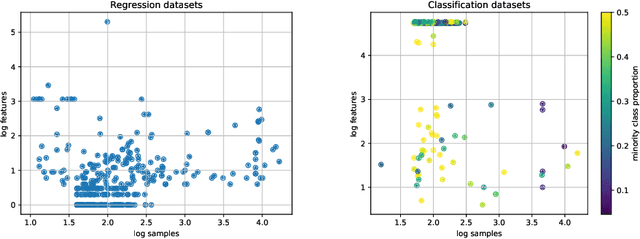
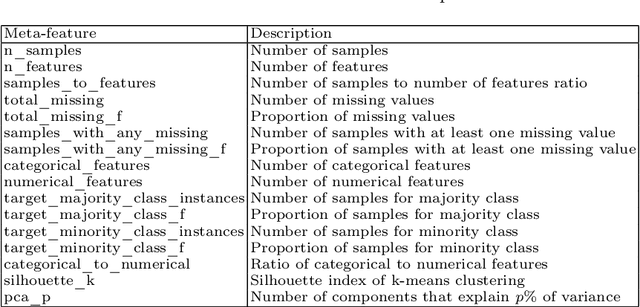
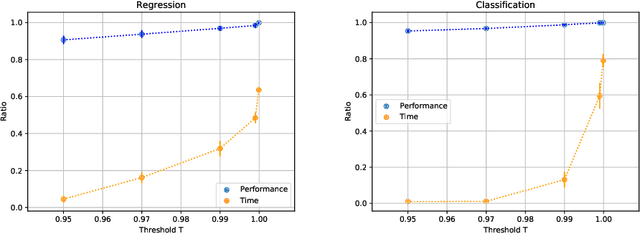
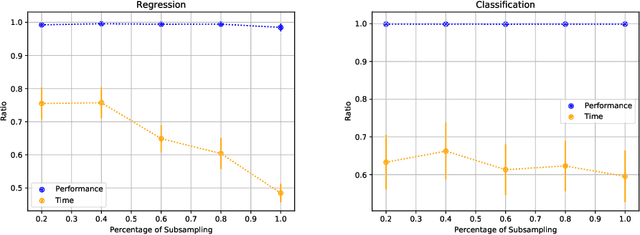
Abstract:AutoML platforms have numerous options for the algorithms to try for each step of the analysis, i.e., different possible algorithms for imputation, transformations, feature selection, and modelling. Finding the optimal combination of algorithms and hyper-parameter values is computationally expensive, as the number of combinations to explore leads to an exponential explosion of the space. In this paper, we present the Sequential Hyper-parameter Space Reduction (SHSR) algorithm that reduces the space for an AutoML tool with negligible drop in its predictive performance. SHSR is a meta-level learning algorithm that analyzes past runs of an AutoML tool on several datasets and learns which hyper-parameter values to filter out from consideration on a new dataset to analyze. SHSR is evaluated on 284 classification and 375 regression problems, showing an approximate 30% reduction in execution time with a performance drop of less than 0.1%.
A Meta-level Analysis of Online Anomaly Detectors
Sep 13, 2022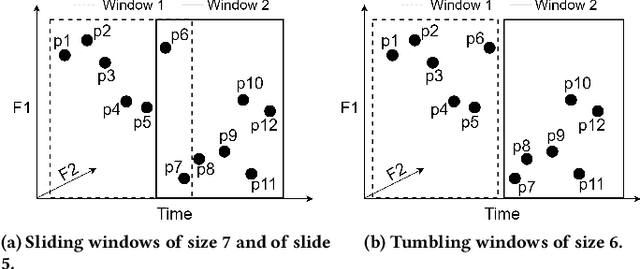
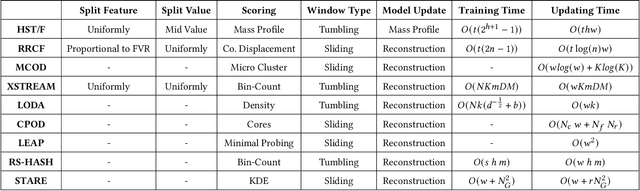
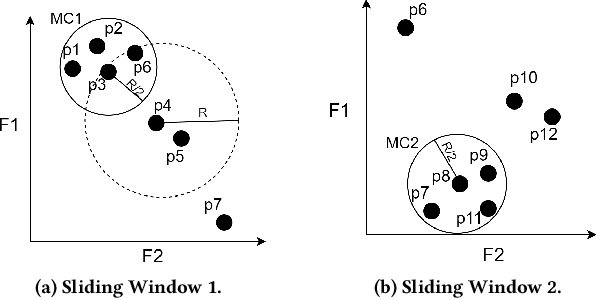
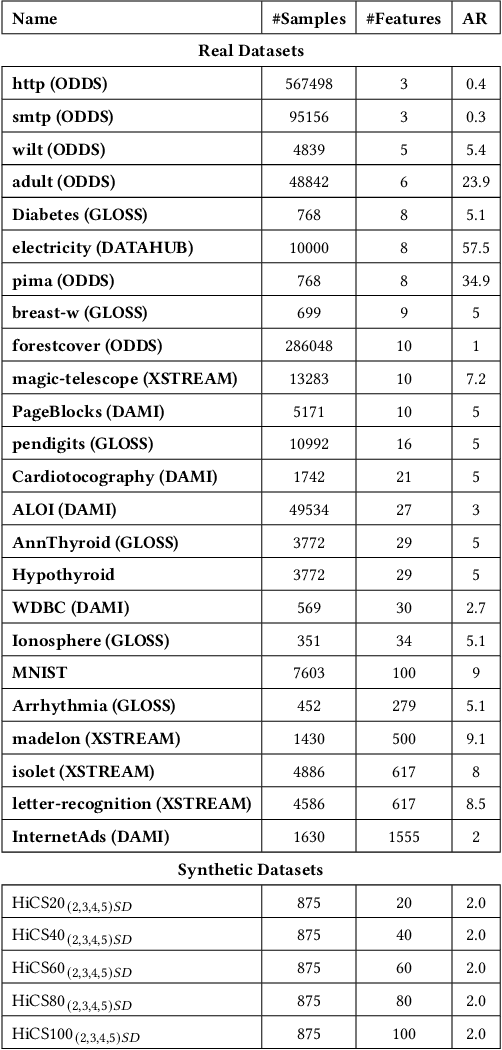
Abstract:Real-time detection of anomalies in streaming data is receiving increasing attention as it allows us to raise alerts, predict faults, and detect intrusions or threats across industries. Yet, little attention has been given to compare the effectiveness and efficiency of anomaly detectors for streaming data (i.e., of online algorithms). In this paper, we present a qualitative, synthetic overview of major online detectors from different algorithmic families (i.e., distance, density, tree or projection-based) and highlight their main ideas for constructing, updating and testing detection models. Then, we provide a thorough analysis of the results of a quantitative experimental evaluation of online detection algorithms along with their offline counterparts. The behavior of the detectors is correlated with the characteristics of different datasets (i.e., meta-features), thereby providing a meta-level analysis of their performance. Our study addresses several missing insights from the literature such as (a) how reliable are detectors against a random classifier and what dataset characteristics make them perform randomly; (b) to what extent online detectors approximate the performance of offline counterparts; (c) which sketch strategy and update primitives of detectors are best to detect anomalies visible only within a feature subspace of a dataset; (d) what are the tradeoffs between the effectiveness and the efficiency of detectors belonging to different algorithmic families; (e) which specific characteristics of datasets yield an online algorithm to outperform all others.
On Predictive Explanation of Data Anomalies
Oct 18, 2021Abstract:Numerous algorithms have been proposed for detecting anomalies (outliers, novelties) in an unsupervised manner. Unfortunately, it is not trivial, in general, to understand why a given sample (record) is labelled as an anomaly and thus diagnose its root causes. We propose the following reduced-dimensionality, surrogate model approach to explain detector decisions: approximate the detection model with another one that employs only a small subset of features. Subsequently, samples can be visualized in this low-dimensionality space for human understanding. To this end, we develop PROTEUS, an AutoML pipeline to produce the surrogate model, specifically designed for feature selection on imbalanced datasets. The PROTEUS surrogate model can not only explain the training data, but also the out-of-sample (unseen) data. In other words, PROTEUS produces predictive explanations by approximating the decision surface of an unsupervised detector. PROTEUS is designed to return an accurate estimate of out-of-sample predictive performance to serve as a metric of the quality of the approximation. Computational experiments confirm the efficacy of PROTEUS to produce predictive explanations for different families of detectors and to reliably estimate their predictive performance in unseen data. Unlike several ad-hoc feature importance methods, PROTEUS is robust to high-dimensional data.
Inference of Stochastic Dynamical Systems from Cross-Sectional Population Data
Dec 09, 2020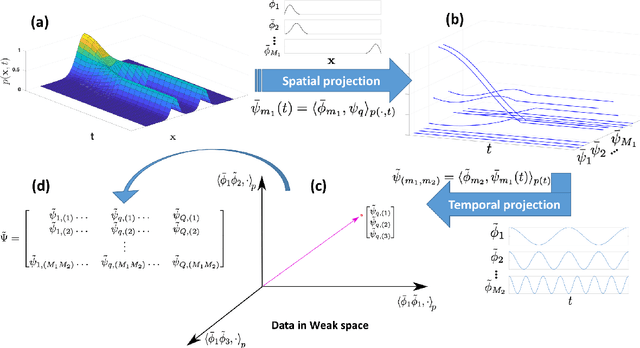
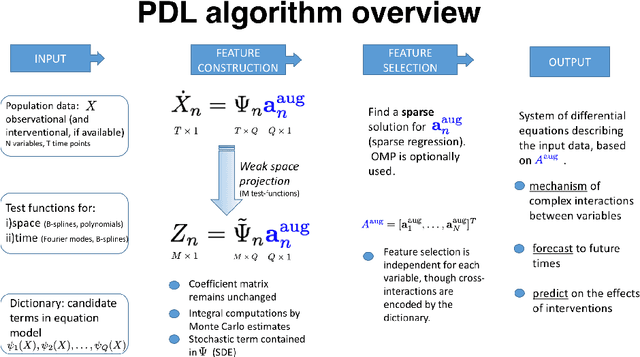
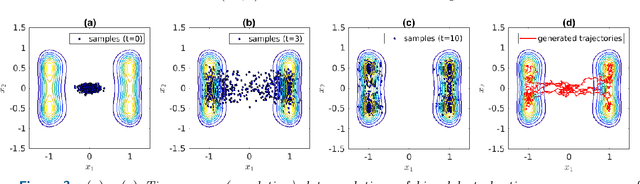
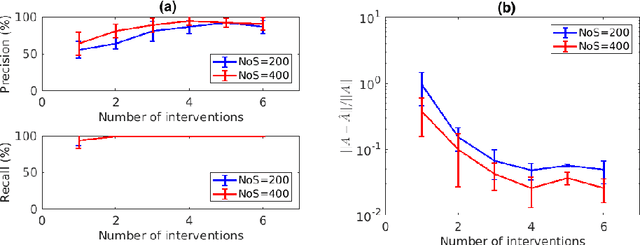
Abstract:Inferring the driving equations of a dynamical system from population or time-course data is important in several scientific fields such as biochemistry, epidemiology, financial mathematics and many others. Despite the existence of algorithms that learn the dynamics from trajectorial measurements there are few attempts to infer the dynamical system straight from population data. In this work, we deduce and then computationally estimate the Fokker-Planck equation which describes the evolution of the population's probability density, based on stochastic differential equations. Then, following the USDL approach, we project the Fokker-Planck equation to a proper set of test functions, transforming it into a linear system of equations. Finally, we apply sparse inference methods to solve the latter system and thus induce the driving forces of the dynamical system. Our approach is illustrated in both synthetic and real data including non-linear, multimodal stochastic differential equations, biochemical reaction networks as well as mass cytometry biological measurements.
A generalised OMP algorithm for feature selection with application to gene expression data
Apr 01, 2020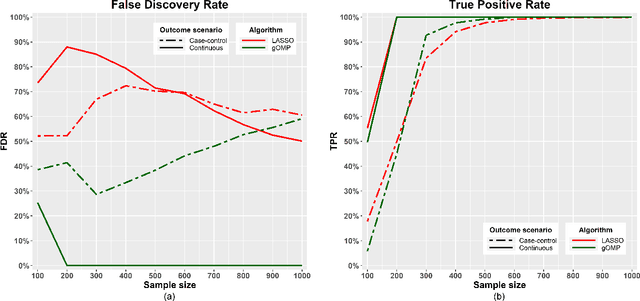
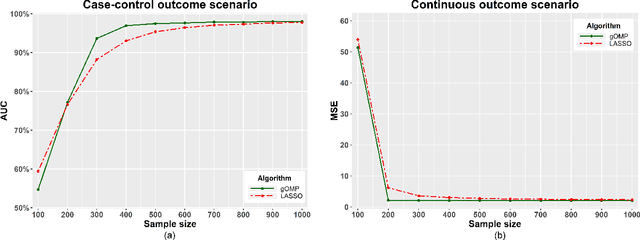
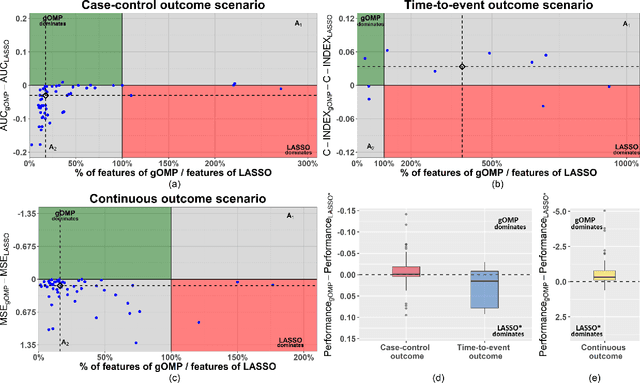
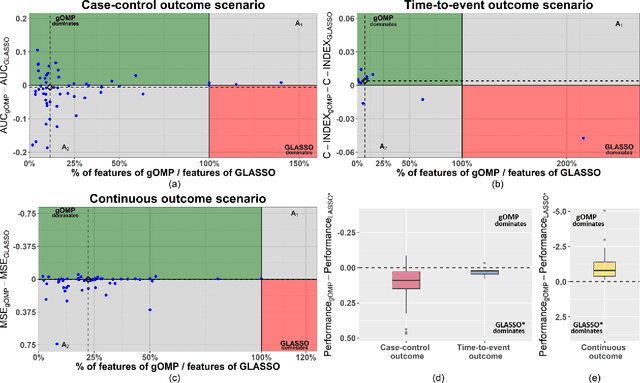
Abstract:Feature selection for predictive analytics is the problem of identifying a minimal-size subset of features that is maximally predictive of an outcome of interest. To apply to molecular data, feature selection algorithms need to be scalable to tens of thousands of available features. In this paper, we propose gOMP, a highly-scalable generalisation of the Orthogonal Matching Pursuit feature selection algorithm to several directions: (a) different types of outcomes, such as continuous, binary, nominal, and time-to-event, (b) different types of predictive models (e.g., linear least squares, logistic regression), (c) different types of predictive features (continuous, categorical), and (d) different, statistical-based stopping criteria. We compare the proposed algorithm against LASSO, a prototypical, widely used algorithm for high-dimensional data. On dozens of simulated datasets, as well as, real gene expression datasets, gOMP is on par, or outperforms LASSO for case-control binary classification, quantified outcomes (regression), and (censored) survival times (time-to-event) analysis. gOMP has also several theoretical advantages that are discussed. While gOMP is based on quite simple and basic statistical ideas, easy to implement and to generalize, we also show in an extensive evaluation that it is also quite effective in bioinformatics analysis settings.
Bootstrapping the Out-of-sample Predictions for Efficient and Accurate Cross-Validation
Aug 25, 2017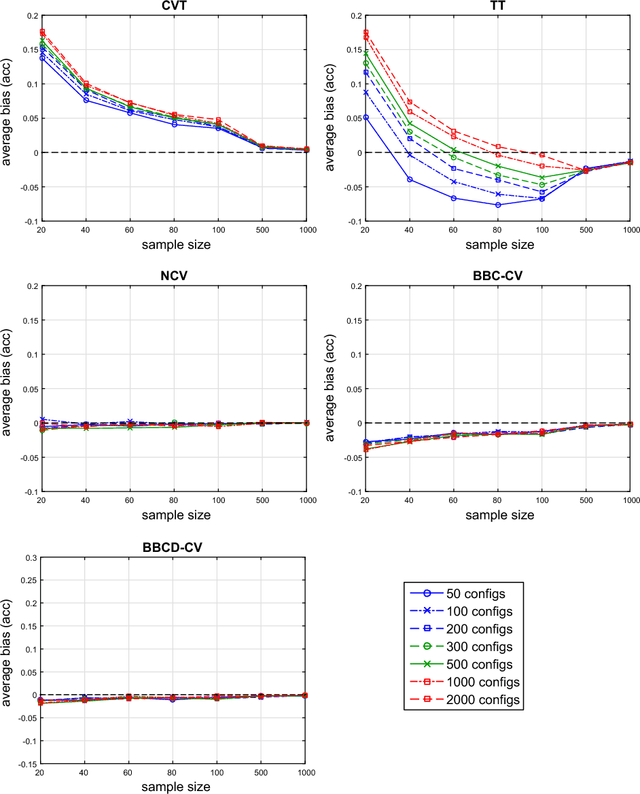
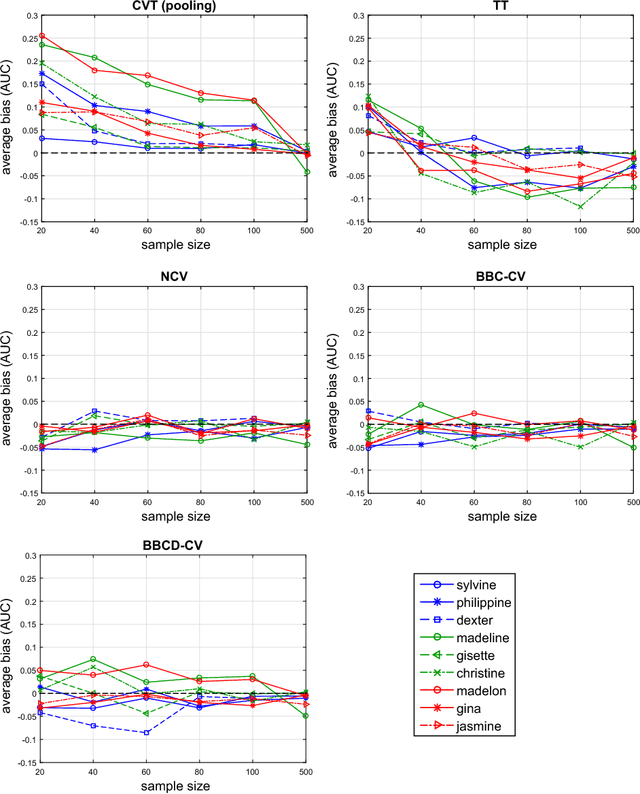
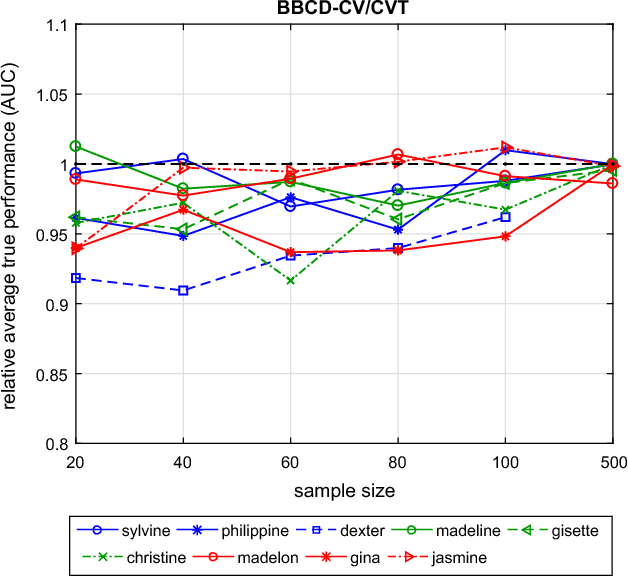
Abstract:Cross-Validation (CV), and out-of-sample performance-estimation protocols in general, are often employed both for (a) selecting the optimal combination of algorithms and values of hyper-parameters (called a configuration) for producing the final predictive model, and (b) estimating the predictive performance of the final model. However, the cross-validated performance of the best configuration is optimistically biased. We present an efficient bootstrap method that corrects for the bias, called Bootstrap Bias Corrected CV (BBC-CV). BBC-CV's main idea is to bootstrap the whole process of selecting the best-performing configuration on the out-of-sample predictions of each configuration, without additional training of models. In comparison to the alternatives, namely the nested cross-validation and a method by Tibshirani and Tibshirani, BBC-CV is computationally more efficient, has smaller variance and bias, and is applicable to any metric of performance (accuracy, AUC, concordance index, mean squared error). Subsequently, we employ again the idea of bootstrapping the out-of-sample predictions to speed up the CV process. Specifically, using a bootstrap-based hypothesis test we stop training of models on new folds of statistically-significantly inferior configurations. We name the method Bootstrap Corrected with Early Dropping CV (BCED-CV) that is both efficient and provides accurate performance estimates.
Massively-Parallel Feature Selection for Big Data
Aug 23, 2017
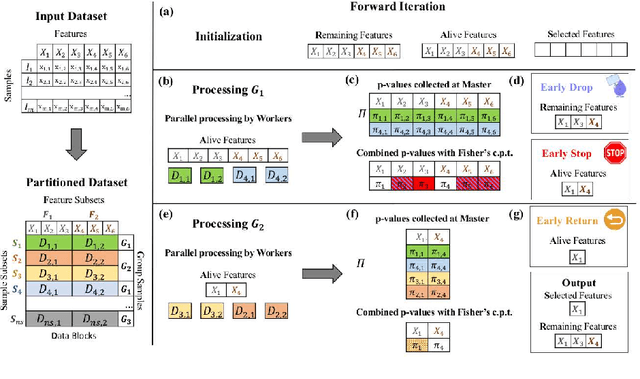
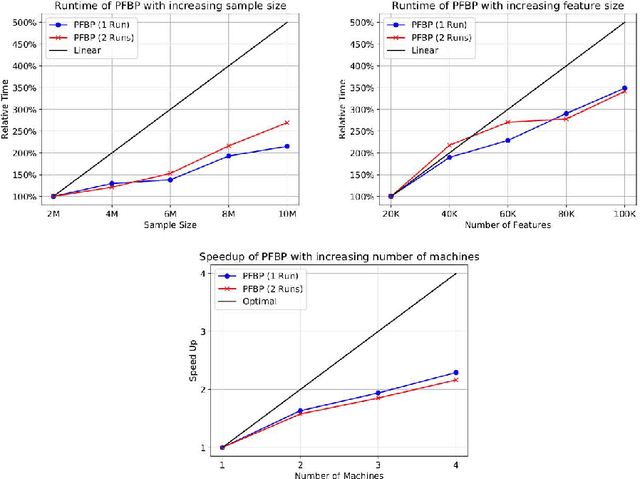
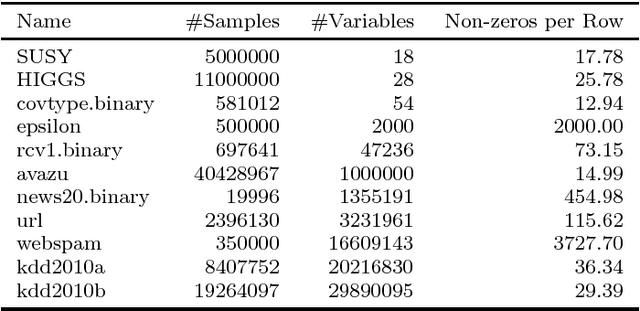
Abstract:We present the Parallel, Forward-Backward with Pruning (PFBP) algorithm for feature selection (FS) in Big Data settings (high dimensionality and/or sample size). To tackle the challenges of Big Data FS PFBP partitions the data matrix both in terms of rows (samples, training examples) as well as columns (features). By employing the concepts of $p$-values of conditional independence tests and meta-analysis techniques PFBP manages to rely only on computations local to a partition while minimizing communication costs. Then, it employs powerful and safe (asymptotically sound) heuristics to make early, approximate decisions, such as Early Dropping of features from consideration in subsequent iterations, Early Stopping of consideration of features within the same iteration, or Early Return of the winner in each iteration. PFBP provides asymptotic guarantees of optimality for data distributions faithfully representable by a causal network (Bayesian network or maximal ancestral graph). Our empirical analysis confirms a super-linear speedup of the algorithm with increasing sample size, linear scalability with respect to the number of features and processing cores, while dominating other competitive algorithms in its class.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge