Inha Kang
3D-Aware Vision-Language Models Fine-Tuning with Geometric Distillation
Jun 11, 2025Abstract:Vision-Language Models (VLMs) have shown remarkable performance on diverse visual and linguistic tasks, yet they remain fundamentally limited in their understanding of 3D spatial structures. We propose Geometric Distillation, a lightweight, annotation-free fine-tuning framework that injects human-inspired geometric cues into pretrained VLMs without modifying their architecture. By distilling (1) sparse correspondences, (2) relative depth relations, and (3) dense cost volumes from off-the-shelf 3D foundation models (e.g., MASt3R, VGGT), our method shapes representations to be geometry-aware while remaining compatible with natural image-text inputs. Through extensive evaluations on 3D vision-language reasoning and 3D perception benchmarks, our method consistently outperforms prior approaches, achieving improved 3D spatial reasoning with significantly lower computational cost. Our work demonstrates a scalable and efficient path to bridge 2D-trained VLMs with 3D understanding, opening up wider use in spatially grounded multimodal tasks.
No Thing, Nothing: Highlighting Safety-Critical Classes for Robust LiDAR Semantic Segmentation in Adverse Weather
Mar 20, 2025Abstract:Existing domain generalization methods for LiDAR semantic segmentation under adverse weather struggle to accurately predict "things" categories compared to "stuff" categories. In typical driving scenes, "things" categories can be dynamic and associated with higher collision risks, making them crucial for safe navigation and planning. Recognizing the importance of "things" categories, we identify their performance drop as a serious bottleneck in existing approaches. We observed that adverse weather induces degradation of semantic-level features and both corruption of local features, leading to a misprediction of "things" as "stuff". To mitigate these corruptions, we suggest our method, NTN - segmeNt Things for No-accident. To address semantic-level feature corruption, we bind each point feature to its superclass, preventing the misprediction of things classes into visually dissimilar categories. Additionally, to enhance robustness against local corruption caused by adverse weather, we define each LiDAR beam as a local region and propose a regularization term that aligns the clean data with its corrupted counterpart in feature space. NTN achieves state-of-the-art performance with a +2.6 mIoU gain on the SemanticKITTI-to-SemanticSTF benchmark and +7.9 mIoU on the SemanticPOSS-to-SemanticSTF benchmark. Notably, NTN achieves a +4.8 and +7.9 mIoU improvement on "things" classes, respectively, highlighting its effectiveness.
Why is the winner the best?
Mar 30, 2023
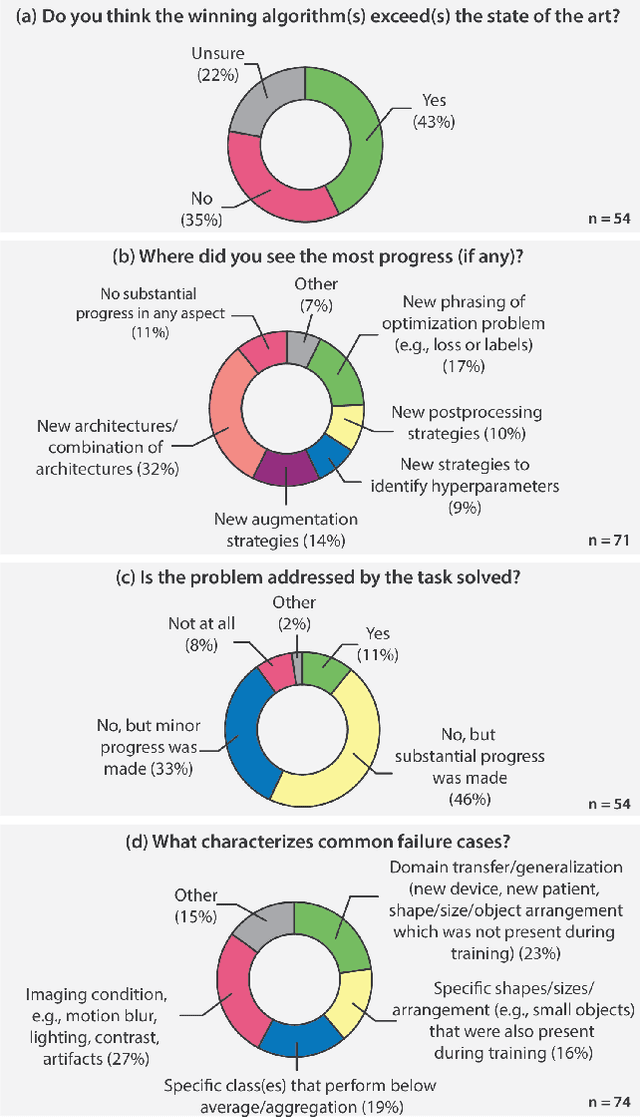
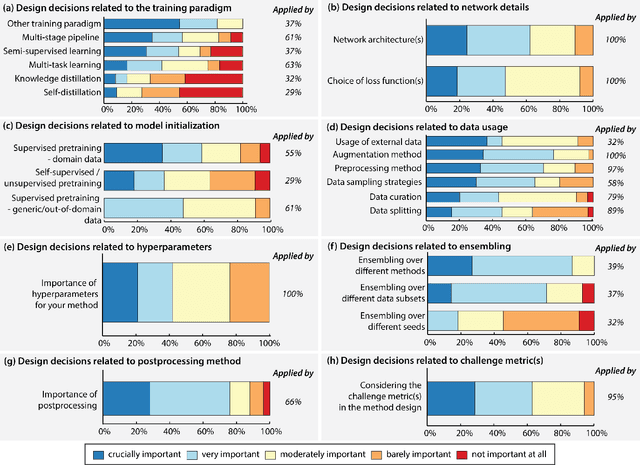
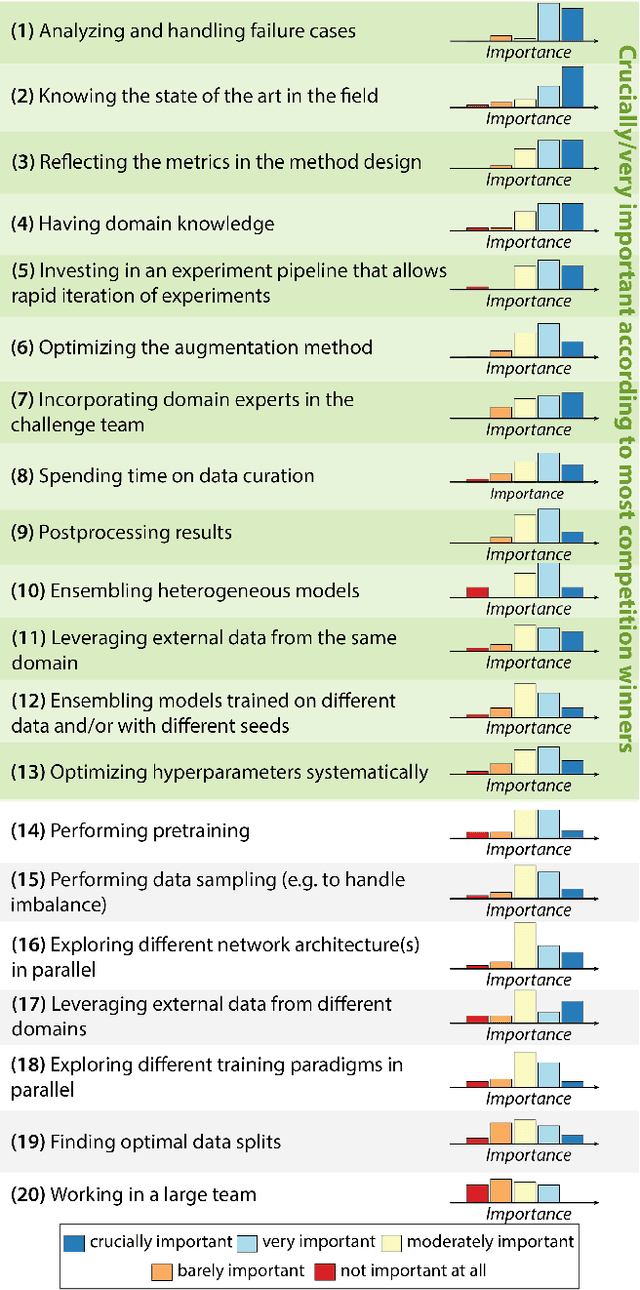
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Joint Embedding of 2D and 3D Networks for Medical Image Anomaly Detection
Dec 23, 2022



Abstract:Obtaining ground truth data in medical imaging has difficulties due to the fact that it requires a lot of annotating time from the experts in the field. Also, when trained with supervised learning, it detects only the cases included in the labels. In real practice, we want to also open to other possibilities than the named cases while examining the medical images. As a solution, the need for anomaly detection that can detect and localize abnormalities by learning the normal characteristics using only normal images is emerging. With medical image data, we can design either 2D or 3D networks of self-supervised learning for anomaly detection task. Although 3D networks, which learns 3D structures of the human body, show good performance in 3D medical image anomaly detection, they cannot be stacked in deeper layers due to memory problems. While 2D networks have advantage in feature detection, they lack 3D context information. In this paper, we develop a method for combining the strength of the 3D network and the strength of the 2D network through joint embedding. We also propose the pretask of self-supervised learning to make it possible for the networks to learn efficiently. Through the experiments, we show that the proposed method achieves better performance in both classification and segmentation tasks compared to the SoTA method.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge