Hanbo Chen
Human-machine Interactive Tissue Prototype Learning for Label-efficient Histopathology Image Segmentation
Nov 26, 2022Abstract:Recently, deep neural networks have greatly advanced histopathology image segmentation but usually require abundant annotated data. However, due to the gigapixel scale of whole slide images and pathologists' heavy daily workload, obtaining pixel-level labels for supervised learning in clinical practice is often infeasible. Alternatively, weakly-supervised segmentation methods have been explored with less laborious image-level labels, but their performance is unsatisfactory due to the lack of dense supervision. Inspired by the recent success of self-supervised learning methods, we present a label-efficient tissue prototype dictionary building pipeline and propose to use the obtained prototypes to guide histopathology image segmentation. Particularly, taking advantage of self-supervised contrastive learning, an encoder is trained to project the unlabeled histopathology image patches into a discriminative embedding space where these patches are clustered to identify the tissue prototypes by efficient pathologists' visual examination. Then, the encoder is used to map the images into the embedding space and generate pixel-level pseudo tissue masks by querying the tissue prototype dictionary. Finally, the pseudo masks are used to train a segmentation network with dense supervision for better performance. Experiments on two public datasets demonstrate that our human-machine interactive tissue prototype learning method can achieve comparable segmentation performance as the fully-supervised baselines with less annotation burden and outperform other weakly-supervised methods. Codes will be available upon publication.
ConCL: Concept Contrastive Learning for Dense Prediction Pre-training in Pathology Images
Jul 14, 2022



Abstract:Detectingandsegmentingobjectswithinwholeslideimagesis essential in computational pathology workflow. Self-supervised learning (SSL) is appealing to such annotation-heavy tasks. Despite the extensive benchmarks in natural images for dense tasks, such studies are, unfortunately, absent in current works for pathology. Our paper intends to narrow this gap. We first benchmark representative SSL methods for dense prediction tasks in pathology images. Then, we propose concept contrastive learning (ConCL), an SSL framework for dense pre-training. We explore how ConCL performs with concepts provided by different sources and end up with proposing a simple dependency-free concept generating method that does not rely on external segmentation algorithms or saliency detection models. Extensive experiments demonstrate the superiority of ConCL over previous state-of-the-art SSL methods across different settings. Along our exploration, we distll several important and intriguing components contributing to the success of dense pre-training for pathology images. We hope this work could provide useful data points and encourage the community to conduct ConCL pre-training for problems of interest. Code is available.
ReMix: A General and Efficient Framework for Multiple Instance Learning based Whole Slide Image Classification
Jul 05, 2022



Abstract:Whole slide image (WSI) classification often relies on deep weakly supervised multiple instance learning (MIL) methods to handle gigapixel resolution images and slide-level labels. Yet the decent performance of deep learning comes from harnessing massive datasets and diverse samples, urging the need for efficient training pipelines for scaling to large datasets and data augmentation techniques for diversifying samples. However, current MIL-based WSI classification pipelines are memory-expensive and computation-inefficient since they usually assemble tens of thousands of patches as bags for computation. On the other hand, despite their popularity in other tasks, data augmentations are unexplored for WSI MIL frameworks. To address them, we propose ReMix, a general and efficient framework for MIL based WSI classification. It comprises two steps: reduce and mix. First, it reduces the number of instances in WSI bags by substituting instances with instance prototypes, i.e., patch cluster centroids. Then, we propose a ``Mix-the-bag'' augmentation that contains four online, stochastic and flexible latent space augmentations. It brings diverse and reliable class-identity-preserving semantic changes in the latent space while enforcing semantic-perturbation invariance. We evaluate ReMix on two public datasets with two state-of-the-art MIL methods. In our experiments, consistent improvements in precision, accuracy, and recall have been achieved but with orders of magnitude reduced training time and memory consumption, demonstrating ReMix's effectiveness and efficiency. Code is available.
Seeking Common Ground While Reserving Differences: Multiple Anatomy Collaborative Framework for Undersampled MRI Reconstruction
Jun 16, 2022


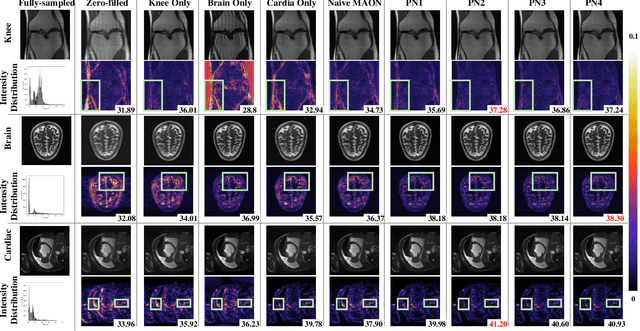
Abstract:Recently, deep neural networks have greatly advanced undersampled Magnetic Resonance Image (MRI) reconstruction, wherein most studies follow the one-anatomy-one-network fashion, i.e., each expert network is trained and evaluated for a specific anatomy. Apart from inefficiency in training multiple independent models, such convention ignores the shared de-aliasing knowledge across various anatomies which can benefit each other. To explore the shared knowledge, one naive way is to combine all the data from various anatomies to train an all-round network. Unfortunately, despite the existence of the shared de-aliasing knowledge, we reveal that the exclusive knowledge across different anatomies can deteriorate specific reconstruction targets, yielding overall performance degradation. Observing this, in this study, we present a novel deep MRI reconstruction framework with both anatomy-shared and anatomy-specific parameterized learners, aiming to "seek common ground while reserving differences" across different anatomies.Particularly, the primary anatomy-shared learners are exposed to different anatomies to model flourishing shared knowledge, while the efficient anatomy-specific learners are trained with their target anatomy for exclusive knowledge. Four different implementations of anatomy-specific learners are presented and explored on the top of our framework in two MRI reconstruction networks. Comprehensive experiments on brain, knee and cardiac MRI datasets demonstrate that three of these learners are able to enhance reconstruction performance via multiple anatomy collaborative learning.
Towards better understanding and better generalization of few-shot classification in histology images with contrastive learning
Feb 18, 2022



Abstract:Few-shot learning is an established topic in natural images for years, but few work is attended to histology images, which is of high clinical value since well-labeled datasets and rare abnormal samples are expensive to collect. Here, we facilitate the study of few-shot learning in histology images by setting up three cross-domain tasks that simulate real clinics problems. To enable label-efficient learning and better generalizability, we propose to incorporate contrastive learning (CL) with latent augmentation (LA) to build a few-shot system. CL learns useful representations without manual labels, while LA transfers semantic variations of the base dataset in an unsupervised way. These two components fully exploit unlabeled training data and can scale gracefully to other label-hungry problems. In experiments, we find i) models learned by CL generalize better than supervised learning for histology images in unseen classes, and ii) LA brings consistent gains over baselines. Prior studies of self-supervised learning mainly focus on ImageNet-like images, which only present a dominant object in their centers. Recent attention has been paid to images with multi-objects and multi-textures. Histology images are a natural choice for such a study. We show the superiority of CL over supervised learning in terms of generalization for such data and provide our empirical understanding for this observation. The findings in this work could contribute to understanding how the model generalizes in the context of both representation learning and histological image analysis. Code is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge