Gollam Rabby
Towards AI-Supported Research: a Vision of the TIB AIssistant
Dec 18, 2025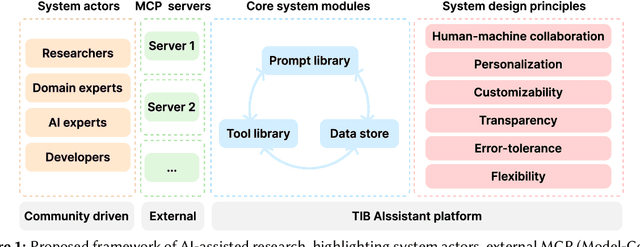
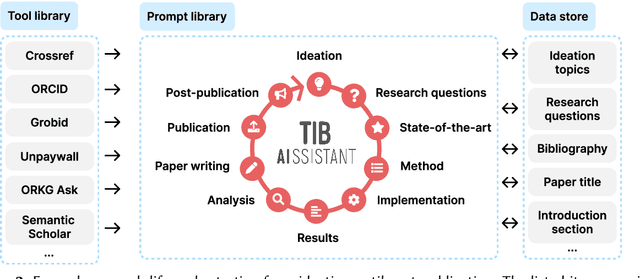
Abstract:The rapid advancements in Generative AI and Large Language Models promise to transform the way research is conducted, potentially offering unprecedented opportunities to augment scholarly workflows. However, effectively integrating AI into research remains a challenge due to varying domain requirements, limited AI literacy, the complexity of coordinating tools and agents, and the unclear accuracy of Generative AI in research. We present the vision of the TIB AIssistant, a domain-agnostic human-machine collaborative platform designed to support researchers across disciplines in scientific discovery, with AI assistants supporting tasks across the research life cycle. The platform offers modular components - including prompt and tool libraries, a shared data store, and a flexible orchestration framework - that collectively facilitate ideation, literature analysis, methodology development, data analysis, and scholarly writing. We describe the conceptual framework, system architecture, and implementation of an early prototype that demonstrates the feasibility and potential impact of our approach.
SLEEPING-DISCO 9M: A large-scale pre-training dataset for generative music modeling
Jun 17, 2025Abstract:We present Sleeping-DISCO 9M, a large-scale pre-training dataset for music and song. To the best of our knowledge, there are no open-source high-quality dataset representing popular and well-known songs for generative music modeling tasks such as text-music, music-captioning, singing-voice synthesis, melody reconstruction and cross-model retrieval. Past contributions focused on isolated and constrained factors whose core perspective was to create synthetic or re-recorded music corpus (e.g. GTSinger, M4Singer) and arbitrarily large-scale audio datasets (e.g. DISCO-10M and LAIONDISCO-12M) had been another focus for the community. Unfortunately, adoption of these datasets has been below substantial in the generative music community as these datasets fail to reflect real-world music and its flavour. Our dataset changes this narrative and provides a dataset that is constructed using actual popular music and world-renowned artists.
EmoNet-Voice: A Fine-Grained, Expert-Verified Benchmark for Speech Emotion Detection
Jun 11, 2025Abstract:The advancement of text-to-speech and audio generation models necessitates robust benchmarks for evaluating the emotional understanding capabilities of AI systems. Current speech emotion recognition (SER) datasets often exhibit limitations in emotional granularity, privacy concerns, or reliance on acted portrayals. This paper introduces EmoNet-Voice, a new resource for speech emotion detection, which includes EmoNet-Voice Big, a large-scale pre-training dataset (featuring over 4,500 hours of speech across 11 voices, 40 emotions, and 4 languages), and EmoNet-Voice Bench, a novel benchmark dataset with human expert annotations. EmoNet-Voice is designed to evaluate SER models on a fine-grained spectrum of 40 emotion categories with different levels of intensities. Leveraging state-of-the-art voice generation, we curated synthetic audio snippets simulating actors portraying scenes designed to evoke specific emotions. Crucially, we conducted rigorous validation by psychology experts who assigned perceived intensity labels. This synthetic, privacy-preserving approach allows for the inclusion of sensitive emotional states often absent in existing datasets. Lastly, we introduce Empathic Insight Voice models that set a new standard in speech emotion recognition with high agreement with human experts. Our evaluations across the current model landscape exhibit valuable findings, such as high-arousal emotions like anger being much easier to detect than low-arousal states like concentration.
EmoNet-Face: An Expert-Annotated Benchmark for Synthetic Emotion Recognition
May 26, 2025Abstract:Effective human-AI interaction relies on AI's ability to accurately perceive and interpret human emotions. Current benchmarks for vision and vision-language models are severely limited, offering a narrow emotional spectrum that overlooks nuanced states (e.g., bitterness, intoxication) and fails to distinguish subtle differences between related feelings (e.g., shame vs. embarrassment). Existing datasets also often use uncontrolled imagery with occluded faces and lack demographic diversity, risking significant bias. To address these critical gaps, we introduce EmoNet Face, a comprehensive benchmark suite. EmoNet Face features: (1) A novel 40-category emotion taxonomy, meticulously derived from foundational research to capture finer details of human emotional experiences. (2) Three large-scale, AI-generated datasets (EmoNet HQ, Binary, and Big) with explicit, full-face expressions and controlled demographic balance across ethnicity, age, and gender. (3) Rigorous, multi-expert annotations for training and high-fidelity evaluation. (4) We build Empathic Insight Face, a model achieving human-expert-level performance on our benchmark. The publicly released EmoNet Face suite - taxonomy, datasets, and model - provides a robust foundation for developing and evaluating AI systems with a deeper understanding of human emotions.
SCI-IDEA: Context-Aware Scientific Ideation Using Token and Sentence Embeddings
Mar 25, 2025



Abstract:Every scientific discovery starts with an idea inspired by prior work, interdisciplinary concepts, and emerging challenges. Recent advancements in large language models (LLMs) trained on scientific corpora have driven interest in AI-supported idea generation. However, generating context-aware, high-quality, and innovative ideas remains challenging. We introduce SCI-IDEA, a framework that uses LLM prompting strategies and Aha Moment detection for iterative idea refinement. SCI-IDEA extracts essential facets from research publications, assessing generated ideas on novelty, excitement, feasibility, and effectiveness. Comprehensive experiments validate SCI-IDEA's effectiveness, achieving average scores of 6.84, 6.86, 6.89, and 6.84 (on a 1-10 scale) across novelty, excitement, feasibility, and effectiveness, respectively. Evaluations employed GPT-4o, GPT-4.5, DeepSeek-32B (each under 2-shot prompting), and DeepSeek-70B (3-shot prompting), with token-level embeddings used for Aha Moment detection. Similarly, it achieves scores of 6.87, 6.86, 6.83, and 6.87 using GPT-4o under 5-shot prompting, GPT-4.5 under 3-shot prompting, DeepSeek-32B under zero-shot chain-of-thought prompting, and DeepSeek-70B under 5-shot prompting with sentence-level embeddings. We also address ethical considerations such as intellectual credit, potential misuse, and balancing human creativity with AI-driven ideation. Our results highlight SCI-IDEA's potential to facilitate the structured and flexible exploration of context-aware scientific ideas, supporting innovation while maintaining ethical standards.
Iterative Hypothesis Generation for Scientific Discovery with Monte Carlo Nash Equilibrium Self-Refining Trees
Mar 25, 2025



Abstract:Scientific hypothesis generation is a fundamentally challenging task in research, requiring the synthesis of novel and empirically grounded insights. Traditional approaches rely on human intuition and domain expertise, while purely large language model (LLM) based methods often struggle to produce hypotheses that are both innovative and reliable. To address these limitations, we propose the Monte Carlo Nash Equilibrium Self-Refine Tree (MC-NEST), a novel framework that integrates Monte Carlo Tree Search with Nash Equilibrium strategies to iteratively refine and validate hypotheses. MC-NEST dynamically balances exploration and exploitation through adaptive sampling strategies, which prioritize high-potential hypotheses while maintaining diversity in the search space. We demonstrate the effectiveness of MC-NEST through comprehensive experiments across multiple domains, including biomedicine, social science, and computer science. MC-NEST achieves average scores of 2.65, 2.74, and 2.80 (on a 1-3 scale) for novelty, clarity, significance, and verifiability metrics on the social science, computer science, and biomedicine datasets, respectively, outperforming state-of-the-art prompt-based methods, which achieve 2.36, 2.51, and 2.52 on the same datasets. These results underscore MC-NEST's ability to generate high-quality, empirically grounded hypotheses across diverse domains. Furthermore, MC-NEST facilitates structured human-AI collaboration, ensuring that LLMs augment human creativity rather than replace it. By addressing key challenges such as iterative refinement and the exploration-exploitation balance, MC-NEST sets a new benchmark in automated hypothesis generation. Additionally, MC-NEST's ethical design enables responsible AI use, emphasizing transparency and human supervision in hypothesis generation.
Project Alexandria: Towards Freeing Scientific Knowledge from Copyright Burdens via LLMs
Feb 26, 2025

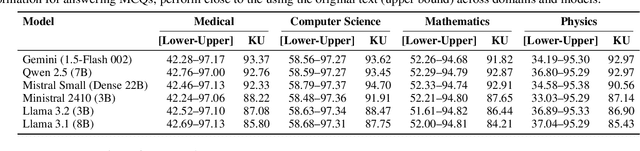

Abstract:Paywalls, licenses and copyright rules often restrict the broad dissemination and reuse of scientific knowledge. We take the position that it is both legally and technically feasible to extract the scientific knowledge in scholarly texts. Current methods, like text embeddings, fail to reliably preserve factual content, and simple paraphrasing may not be legally sound. We urge the community to adopt a new idea: convert scholarly documents into Knowledge Units using LLMs. These units use structured data capturing entities, attributes and relationships without stylistic content. We provide evidence that Knowledge Units: (1) form a legally defensible framework for sharing knowledge from copyrighted research texts, based on legal analyses of German copyright law and U.S. Fair Use doctrine, and (2) preserve most (~95%) factual knowledge from original text, measured by MCQ performance on facts from the original copyrighted text across four research domains. Freeing scientific knowledge from copyright promises transformative benefits for scientific research and education by allowing language models to reuse important facts from copyrighted text. To support this, we share open-source tools for converting research documents into Knowledge Units. Overall, our work posits the feasibility of democratizing access to scientific knowledge while respecting copyright.
SelfCheckAgent: Zero-Resource Hallucination Detection in Generative Large Language Models
Feb 03, 2025Abstract:Detecting hallucinations in Large Language Models (LLMs) remains a critical challenge for their reliable deployment in real-world applications. To address this, we introduce SelfCheckAgent, a novel framework integrating three different agents: the Symbolic Agent, the Specialized Detection Agent, and the Contextual Consistency Agent. These agents provide a robust multi-dimensional approach to hallucination detection. Notable results include the Contextual Consistency Agent leveraging Llama 3.1 with Chain-of-Thought (CoT) to achieve outstanding performance on the WikiBio dataset, with NonFactual hallucination detection scoring 93.64%, Factual 70.26%, and Ranking 78.48% respectively. On the AIME dataset, GPT-4o with CoT excels in NonFactual detection with 94.89% but reveals trade-offs in Factual with 30.58% and Ranking with 30.68%, underscoring the complexity of hallucination detection in the complex mathematical domains. The framework also incorporates a triangulation strategy, which increases the strengths of the SelfCheckAgent, yielding significant improvements in real-world hallucination identification. The comparative analysis demonstrates SelfCheckAgent's applicability across diverse domains, positioning it as a crucial advancement for trustworthy LLMs. These findings highlight the potentiality of consistency-driven methodologies in detecting hallucinations in LLMs.
MC-NEST -- Enhancing Mathematical Reasoning in Large Language Models with a Monte Carlo Nash Equilibrium Self-Refine Tree
Nov 23, 2024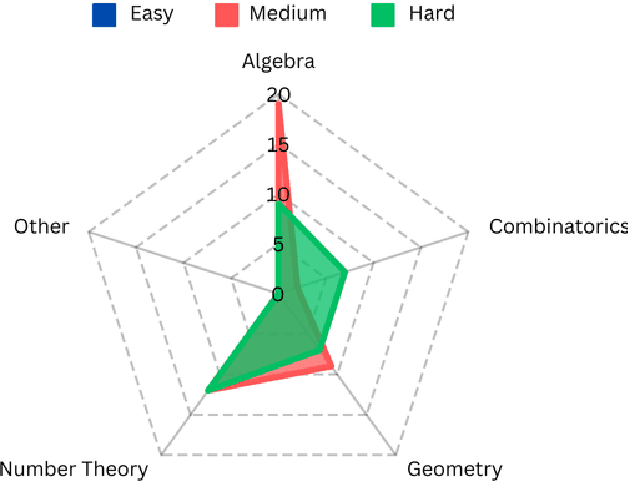

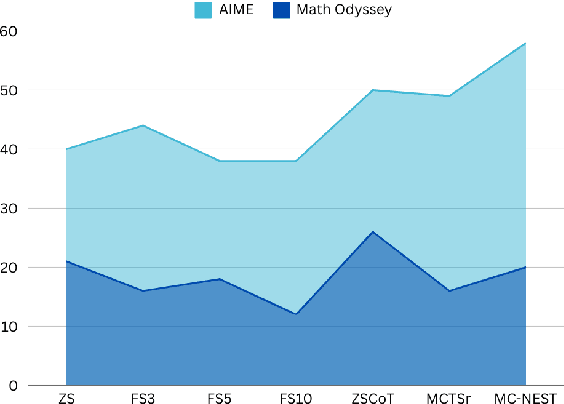

Abstract:Mathematical reasoning has proven to be a critical yet challenging task for large language models (LLMs), as they often struggle with complex multi-step problems. To address these limitations, we introduce the Monte Carlo Nash Equilibrium Self-Refine Tree (MC-NEST) algorithm, an enhancement of the Monte Carlo Tree Self-Refine (MCTSr) approach. By integrating Nash Equilibrium strategies with LLM-based self-refinement and self-evaluation processes, MC-NEST aims to improve decision-making for complex mathematical reasoning tasks. This method ensures balanced exploration and exploitation of potential solutions, leveraging Upper Confidence Bound (UCT) scores and various selection policies. Through iterative critique and refinement, MC-NEST enhances the reasoning capabilities of LLMs, particularly for problems requiring strategic decision-making. Comparative analysis reveals that GPT-4o, equipped with MC-NEST using an Importance Sampling Policy, achieved superior accuracy in domains such as Number Theory and Geometry. These results suggest that both LLMs GPT-4o and Phi-3-mini can benefit from MC-NEST, with iterative self-refinement proving especially effective in expanding the reasoning capacity and problem-solving performance of LLMs. We evaluate the effectiveness of MC-NEST on challenging Olympiad-level benchmarks, demonstrating its potential to significantly boost complex mathematical reasoning performance in LLMs.
NeuroSym-BioCAT: Leveraging Neuro-Symbolic Methods for Biomedical Scholarly Document Categorization and Question Answering
Oct 29, 2024



Abstract:The growing volume of biomedical scholarly document abstracts presents an increasing challenge in efficiently retrieving accurate and relevant information. To address this, we introduce a novel approach that integrates an optimized topic modelling framework, OVB-LDA, with the BI-POP CMA-ES optimization technique for enhanced scholarly document abstract categorization. Complementing this, we employ the distilled MiniLM model, fine-tuned on domain-specific data, for high-precision answer extraction. Our approach is evaluated across three configurations: scholarly document abstract retrieval, gold-standard scholarly documents abstract, and gold-standard snippets, consistently outperforming established methods such as RYGH and bio-answer finder. Notably, we demonstrate that extracting answers from scholarly documents abstracts alone can yield high accuracy, underscoring the sufficiency of abstracts for many biomedical queries. Despite its compact size, MiniLM exhibits competitive performance, challenging the prevailing notion that only large, resource-intensive models can handle such complex tasks. Our results, validated across various question types and evaluation batches, highlight the robustness and adaptability of our method in real-world biomedical applications. While our approach shows promise, we identify challenges in handling complex list-type questions and inconsistencies in evaluation metrics. Future work will focus on refining the topic model with more extensive domain-specific datasets, further optimizing MiniLM and utilizing large language models (LLM) to improve both precision and efficiency in biomedical question answering.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge