Gexin Huang
Interactive Tumor Progression Modeling via Sketch-Based Image Editing
Mar 10, 2025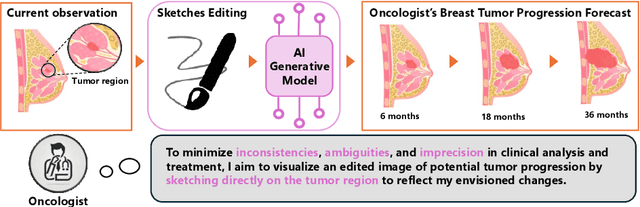
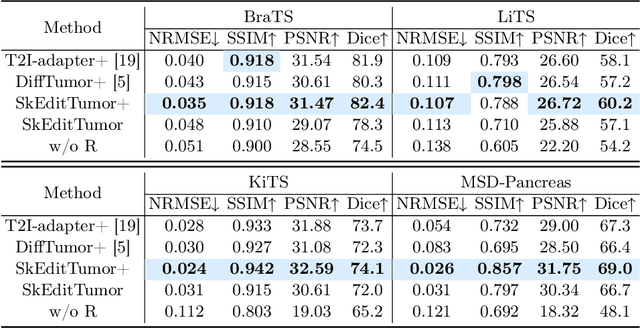
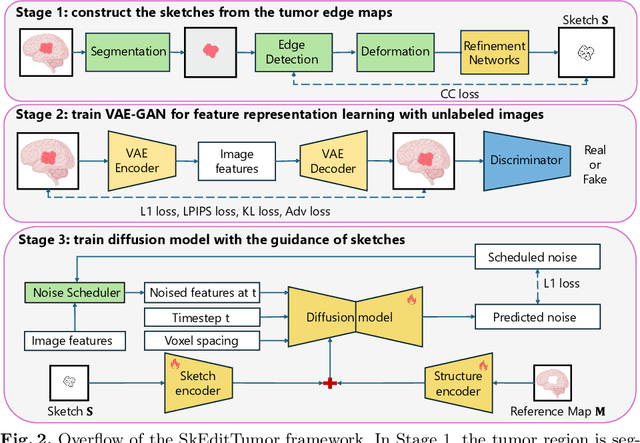
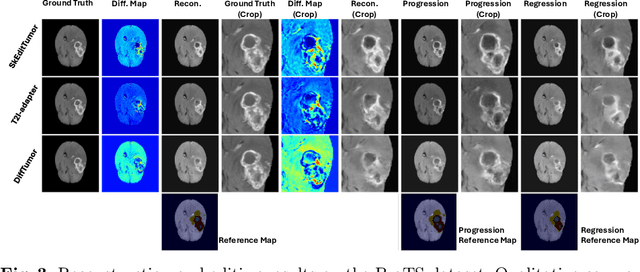
Abstract:Accurately visualizing and editing tumor progression in medical imaging is crucial for diagnosis, treatment planning, and clinical communication. To address the challenges of subjectivity and limited precision in existing methods, we propose SkEditTumor, a sketch-based diffusion model for controllable tumor progression editing. By leveraging sketches as structural priors, our method enables precise modifications of tumor regions while maintaining structural integrity and visual realism. We evaluate SkEditTumor on four public datasets - BraTS, LiTS, KiTS, and MSD-Pancreas - covering diverse organs and imaging modalities. Experimental results demonstrate that our method outperforms state-of-the-art baselines, achieving superior image fidelity and segmentation accuracy. Our contributions include a novel integration of sketches with diffusion models for medical image editing, fine-grained control over tumor progression visualization, and extensive validation across multiple datasets, setting a new benchmark in the field.
Predicting Genetic Mutation from Whole Slide Images via Biomedical-Linguistic Knowledge Enhanced Multi-label Classification
Jun 05, 2024Abstract:Predicting genetic mutations from whole slide images is indispensable for cancer diagnosis. However, existing work training multiple binary classification models faces two challenges: (a) Training multiple binary classifiers is inefficient and would inevitably lead to a class imbalance problem. (b) The biological relationships among genes are overlooked, which limits the prediction performance. To tackle these challenges, we innovatively design a Biological-knowledge enhanced PathGenomic multi-label Transformer to improve genetic mutation prediction performances. BPGT first establishes a novel gene encoder that constructs gene priors by two carefully designed modules: (a) A gene graph whose node features are the genes' linguistic descriptions and the cancer phenotype, with edges modeled by genes' pathway associations and mutation consistencies. (b) A knowledge association module that fuses linguistic and biomedical knowledge into gene priors by transformer-based graph representation learning, capturing the intrinsic relationships between different genes' mutations. BPGT then designs a label decoder that finally performs genetic mutation prediction by two tailored modules: (a) A modality fusion module that firstly fuses the gene priors with critical regions in WSIs and obtains gene-wise mutation logits. (b) A comparative multi-label loss that emphasizes the inherent comparisons among mutation status to enhance the discrimination capabilities. Sufficient experiments on The Cancer Genome Atlas benchmark demonstrate that BPGT outperforms the state-of-the-art.
Electromagnetic Source Imaging via a Data-Synthesis-Based Denoising Autoencoder
Oct 31, 2020



Abstract:Electromagnetic source imaging (ESI) is a highly ill-posed inverse problem. To find a unique solution, traditional ESI methods impose a variety of priors that may not reflect the actual source properties. Such limitations of traditional ESI methods hinder their further applications. Inspired by deep learning approaches, a novel data-synthesized spatio-temporal denoising autoencoder method (DST-DAE) method was proposed to solve the ESI inverse problem. Unlike the traditional methods, we utilize a neural network to directly seek generalized mapping from the measured E/MEG signals to the cortical sources. A novel data synthesis strategy is employed by introducing the prior information of sources to the generated large-scale samples using the forward model of ESI. All the generated data are used to drive the neural network to automatically learn inverse mapping. To achieve better estimation performance, a denoising autoencoder (DAE) architecture with spatio-temporal feature extraction blocks is designed. Compared with the traditional methods, we show (1) that the novel deep learning approach provides an effective and easy-to-apply way to solve the ESI problem, that (2) compared to traditional methods, DST-DAE with the data synthesis strategy can better consider the characteristics of real sources than the mathematical formulation of prior assumptions, and that (3) the specifically designed architecture of DAE can not only provide a better estimation of source signals but also be robust to noise pollution. Extensive numerical experiments show that the proposed method is superior to the traditional knowledge-driven ESI methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge