Gabriella Pizzuto
MATTERIX: toward a digital twin for robotics-assisted chemistry laboratory automation
Jan 19, 2026Abstract:Accelerated materials discovery is critical for addressing global challenges. However, developing new laboratory workflows relies heavily on real-world experimental trials, and this can hinder scalability because of the need for numerous physical make-and-test iterations. Here we present MATTERIX, a multiscale, graphics processing unit-accelerated robotic simulation framework designed to create high-fidelity digital twins of chemistry laboratories, thus accelerating workflow development. This multiscale digital twin simulates robotic physical manipulation, powder and liquid dynamics, device functionalities, heat transfer and basic chemical reaction kinetics. This is enabled by integrating realistic physics simulation and photorealistic rendering with a modular graphics processing unit-accelerated semantics engine, which models logical states and continuous behaviors to simulate chemistry workflows across different levels of abstraction. MATTERIX streamlines the creation of digital twin environments through open-source asset libraries and interfaces, while enabling flexible workflow design via hierarchical plan definition and a modular skill library that incorporates learning-based methods. Our approach demonstrates sim-to-real transfer in robotic chemistry setups, reducing reliance on costly real-world experiments and enabling the testing of hypothetical automated workflows in silico. The project website is available at https://accelerationconsortium.github.io/Matterix/ .
FLIP: Flowability-Informed Powder Weighing
Jun 05, 2025Abstract:Autonomous manipulation of powders remains a significant challenge for robotic automation in scientific laboratories. The inherent variability and complex physical interactions of powders in flow, coupled with variability in laboratory conditions necessitates adaptive automation. This work introduces FLIP, a flowability-informed powder weighing framework designed to enhance robotic policy learning for granular material handling. Our key contribution lies in using material flowability, quantified by the angle of repose, to optimise physics-based simulations through Bayesian inference. This yields material-specific simulation environments capable of generating accurate training data, which reflects diverse powder behaviours, for training "robot chemists". Building on this, FLIP integrates quantified flowability into a curriculum learning strategy, fostering efficient acquisition of robust robotic policies by gradually introducing more challenging, less flowable powders. We validate the efficacy of our method on a robotic powder weighing task under real-world laboratory conditions. Experimental results show that FLIP with a curriculum strategy achieves a low dispensing error of 2.12 +/- 1.53 mg, outperforming methods that do not leverage flowability data, such as domain randomisation (6.11 +/- 3.92 mg). These results demonstrate FLIP's improved ability to generalise to previously unseen, more cohesive powders and to new target masses.
Accelerating Discovery in Natural Science Laboratories with AI and Robotics: Perspectives and Challenges from the 2024 IEEE ICRA Workshop, Yokohama, Japan
Jan 12, 2025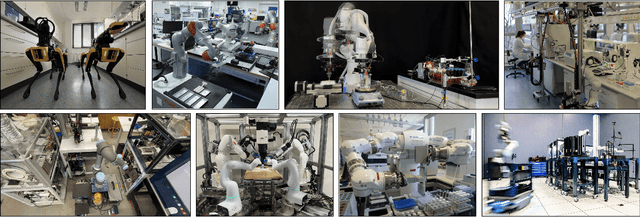
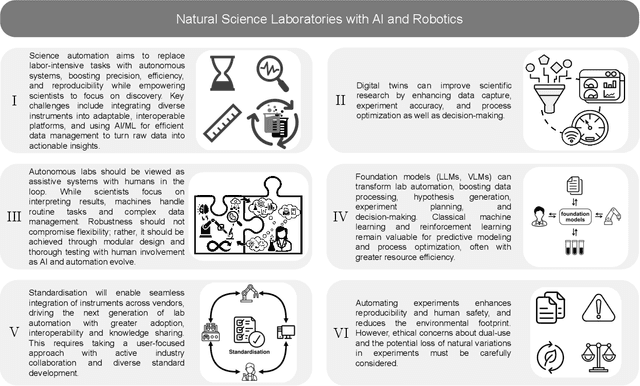
Abstract:Science laboratory automation enables accelerated discovery in life sciences and materials. However, it requires interdisciplinary collaboration to address challenges such as robust and flexible autonomy, reproducibility, throughput, standardization, the role of human scientists, and ethics. This article highlights these issues, reflecting perspectives from leading experts in laboratory automation across different disciplines of the natural sciences.
Powder-Bot: A Modular Autonomous Multi-Robot Workflow for Powder X-Ray Diffraction
Sep 01, 2023Abstract:Powder X-ray diffraction (PXRD) is a key technique for the structural characterisation of solid-state materials, but compared with tasks such as liquid handling, its end-to-end automation is highly challenging. This is because coupling PXRD experiments with crystallisation comprises multiple solid handling steps that include sample recovery, sample preparation by grinding, sample mounting and, finally, collection of X-ray diffraction data. Each of these steps has individual technical challenges from an automation perspective, and hence no commercial instrument exists that can grow crystals, process them into a powder, mount them in a diffractometer, and collect PXRD data in an autonomous, closed-loop way. Here we present an automated robotic workflow to carry out autonomous PXRD experiments. The PXRD data collected for polymorphs of small organic compounds is comparable to that collected under the same conditions manually. Beyond accelerating PXRD experiments, this workflow involves 13 component steps and integrates three different types of robots, each from a separate supplier, illustrating the power of flexible, modular automation in complex, multitask laboratories.
Leveraging Multi-modal Sensing for Robotic Insertion Tasks in R&D Laboratories
Jul 02, 2023Abstract:Performing a large volume of experiments in Chemistry labs creates repetitive actions costing researchers time, automating these routines is highly desirable. Previous experiments in robotic chemistry have performed high numbers of experiments autonomously, however, these processes rely on automated machines in all stages from solid or liquid addition to analysis of the final product. In these systems every transition between machine requires the robotic chemist to pick and place glass vials, however, this is currently performed using open loop methods which require all equipment being used by the robot to be in well defined known locations. We seek to begin closing the loop in this vial handling process in a way which also fosters human-robot collaboration in the chemistry lab environment. To do this the robot must be able to detect valid placement positions for the vials it is collecting, and reliably insert them into the detected locations. We create a single modality visual method for estimating placement locations to provide a baseline before introducing two additional methods of feedback (force and tactile feedback). Our visual method uses a combination of classic computer vision methods and a CNN discriminator to detect possible insertion points, then a vial is grasped and positioned above an insertion point and the multi-modal methods guide the final insertion movements using an efficient search pattern. Through our experiments we show the baseline insertion rate of 48.78% improves to 89.55% with the addition of our "force and vision" multi-modal feedback method.
Accelerating Laboratory Automation Through Robot Skill Learning For Sample Scraping
Sep 29, 2022



Abstract:The potential use of robotics for laboratory experiments offers an attractive route to alleviate scientists from tedious tasks while accelerating the process of obtaining new materials, where topical issues such as climate change and disease risks worldwide would greatly benefit. While some experimental workflows can already benefit from automation, it is common that sample preparation is still carried out manually due to the high level of motor function required when dealing with heterogeneous systems, e.g., different tools, chemicals, and glassware. A fundamental workflow in chemical fields is crystallisation, where one application is polymorph screening, i.e., obtaining a three dimensional molecular structure from a crystal. For this process, it is of utmost importance to recover as much of the sample as possible since synthesising molecules is both costly in time and money. To this aim, chemists have to scrape vials to retrieve sample contents prior to imaging plate transfer. Automating this process is challenging as it goes beyond robotic insertion tasks due to a fundamental requirement of having to execute fine-granular movements within a constrained environment that is the sample vial. Motivated by how human chemists carry out this process of scraping powder from vials, our work proposes a model-free reinforcement learning method for learning a scraping policy, leading to a fully autonomous sample scraping procedure. To realise that, we first create a simulation environment with a Panda Franka Emika robot using a laboratory scraper which is inserted into a simulated vial, to demonstrate how a scraping policy can be learned successfully. We then evaluate our method on a real robotic manipulator in laboratory settings, and show that our method can autonomously scrape powder across various setups.
Efficient Learning of Inverse Dynamics Models for Adaptive Computed Torque Control
May 10, 2022



Abstract:Modelling robot dynamics accurately is essential for control, motion optimisation and safe human-robot collaboration. Given the complexity of modern robotic systems, dynamics modelling remains non-trivial, mostly in the presence of compliant actuators, mechanical inaccuracies, friction and sensor noise. Recent efforts have focused on utilising data-driven methods such as Gaussian processes and neural networks to overcome these challenges, as they are capable of capturing these dynamics without requiring extensive knowledge beforehand. While Gaussian processes have shown to be an effective method for learning robotic dynamics with the ability to also represent the uncertainty in the learned model through its variance, they come at a cost of cubic time complexity rather than linear, as is the case for deep neural networks. In this work, we leverage the use of deep kernel models, which combine the computational efficiency of deep learning with the non-parametric flexibility of kernel methods (Gaussian processes), with the overarching goal of realising an accurate probabilistic framework for uncertainty quantification. Through using the predicted variance, we adapt the feedback gains as more accurate models are learned, leading to low-gain control without compromising tracking accuracy. Using simulated and real data recorded from a seven degree-of-freedom robotic manipulator, we illustrate how using stochastic variational inference with deep kernel models increases compliance in the computed torque controller, and retains tracking accuracy. We empirically show how our model outperforms current state-of-the-art methods with prediction uncertainty for online inverse dynamics model learning, and solidify its adaptation and generalisation capabilities across different setups.
ARChemist: Autonomous Robotic Chemistry System Architecture
Apr 28, 2022



Abstract:Automated laboratory experiments have the potential to propel new discoveries, while increasing reproducibility and improving scientists' safety when handling dangerous materials. However, many automated laboratory workflows have not fully leveraged the remarkable advancements in robotics and digital lab equipment. As a result, most robotic systems used in the labs are programmed specifically for a single experiment, often relying on proprietary architectures or using unconventional hardware. In this work, we tackle this problem by proposing a novel robotic system architecture specifically designed with and for chemists, which allows the scientist to easily reconfigure their setup for new experiments. Specifically, the system's strength is its ability to combine together heterogeneous robotic platforms with standard laboratory equipment to create different experimental setups. Finally, we show how the architecture can be used for specific laboratory experiments through case studies such as solubility screening and crystallisation.
SOLIS: Autonomous Solubility Screening using Deep Neural Networks
Mar 18, 2022



Abstract:Accelerating material discovery has tremendous societal and industrial impact, particularly for pharmaceuticals and clean energy production. Many experimental instruments have some degree of automation, facilitating continuous running and higher throughput. However, it is common that sample preparation is still carried out manually. This can result in researchers spending a significant amount of their time on repetitive tasks, which introduces errors and can prohibit production of statistically relevant data. Crystallisation experiments are common in many chemical fields, both for purification and in polymorph screening experiments. The initial step often involves a solubility screen of the molecule; that is, understanding whether molecular compounds have dissolved in a particular solvent. This usually can be time consuming and work intensive. Moreover, accurate knowledge of the precise solubility limit of the molecule is often not required, and simply measuring a threshold of solubility in each solvent would be sufficient. To address this, we propose a novel cascaded deep model that is inspired by how a human chemist would visually assess a sample to determine whether the solid has completely dissolved in the solution. In this paper, we design, develop, and evaluate the first fully autonomous solubility screening framework, which leverages state-of-the-art methods for image segmentation and convolutional neural networks for image classification. To realise that, we first create a dataset comprising different molecules and solvents, which is collected in a real-world chemistry laboratory. We then evaluated our method on the data recorded through an eye-in-hand camera mounted on a seven degree-of-freedom robotic manipulator, and show that our model can achieve 99.13% test accuracy across various setups.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge