Evrim Acar
dCMF: Learning interpretable evolving patterns from temporal multiway data
Feb 26, 2025


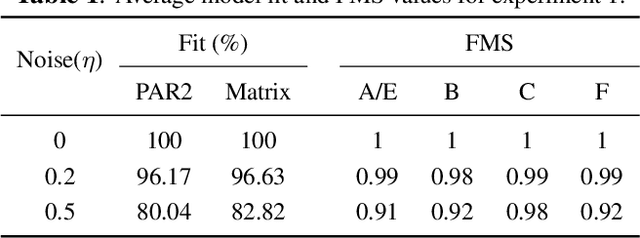
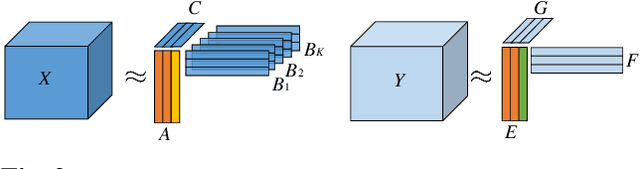
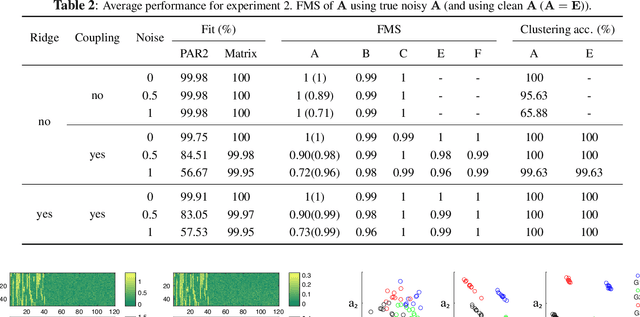
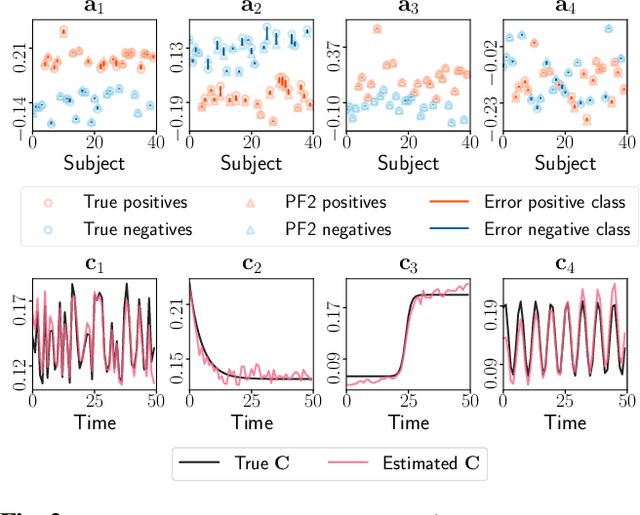
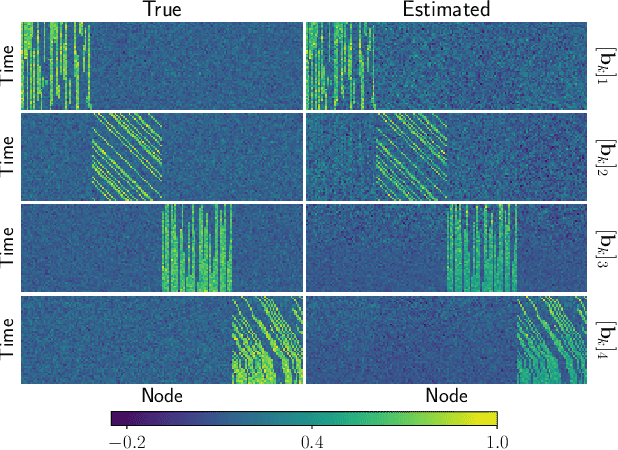
Abstract:Multiway datasets are commonly analyzed using unsupervised matrix and tensor factorization methods to reveal underlying patterns. Frequently, such datasets include timestamps and could correspond to, for example, health-related measurements of subjects collected over time. The temporal dimension is inherently different from the other dimensions, requiring methods that account for its intrinsic properties. Linear Dynamical Systems (LDS) are specifically designed to capture sequential dependencies in the observed data. In this work, we bridge the gap between tensor factorizations and dynamical modeling by exploring the relationship between LDS, Coupled Matrix Factorizations (CMF) and the PARAFAC2 model. We propose a time-aware coupled factorization model called d(ynamical)CMF that constrains the temporal evolution of the latent factors to adhere to a specific LDS structure. Using synthetic datasets, we compare the performance of dCMF with PARAFAC2 and t(emporal)PARAFAC2 which incorporates temporal smoothness. Our results show that dCMF and PARAFAC2-based approaches perform similarly when capturing smoothly evolving patterns that adhere to the PARAFAC2 structure. However, dCMF outperforms alternatives when the patterns evolve smoothly but deviate from the PARAFAC2 structure. Furthermore, we demonstrate that the proposed dCMF method enables to capture more complex dynamics when additional prior information about the temporal evolution is incorporated.
tPARAFAC2: Tracking evolving patterns in (incomplete) temporal data
Jul 01, 2024Abstract:Tensor factorizations have been widely used for the task of uncovering patterns in various domains. Often, the input is time-evolving, shifting the goal to tracking the evolution of underlying patterns instead. To adapt to this more complex setting, existing methods incorporate temporal regularization but they either have overly constrained structural requirements or lack uniqueness which is crucial for interpretation. In this paper, in order to capture the underlying evolving patterns, we introduce t(emporal)PARAFAC2 which utilizes temporal smoothness regularization on the evolving factors. We propose an algorithmic framework that employs Alternating Optimization (AO) and the Alternating Direction Method of Multipliers (ADMM) to fit the model. Furthermore, we extend the algorithmic framework to the case of partially observed data. Our numerical experiments on both simulated and real datasets demonstrate the effectiveness of the temporal smoothness regularization, in particular, in the case of data with missing entries. We also provide an extensive comparison of different approaches for handling missing data within the proposed framework.
A Time-aware tensor decomposition for tracking evolving patterns
Aug 15, 2023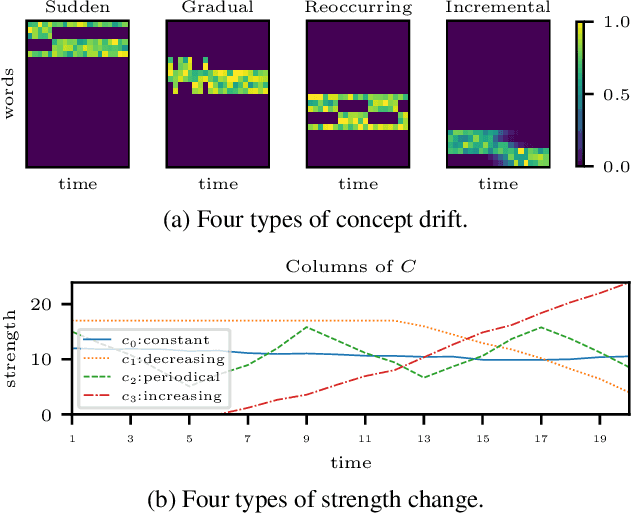
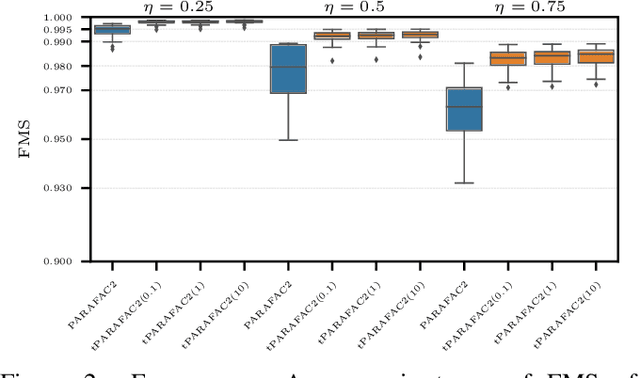
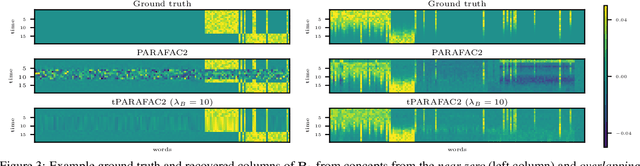
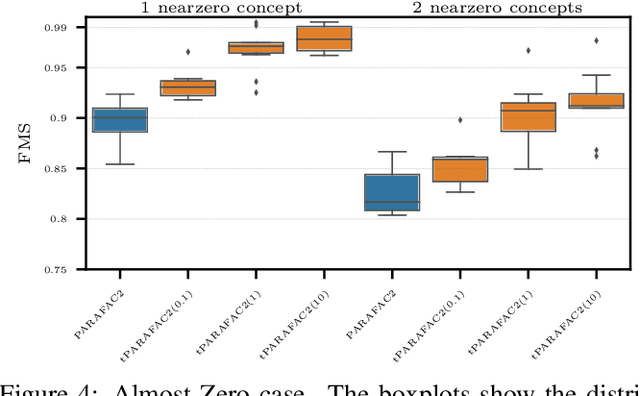
Abstract:Time-evolving data sets can often be arranged as a higher-order tensor with one of the modes being the time mode. While tensor factorizations have been successfully used to capture the underlying patterns in such higher-order data sets, the temporal aspect is often ignored, allowing for the reordering of time points. In recent studies, temporal regularizers are incorporated in the time mode to tackle this issue. Nevertheless, existing approaches still do not allow underlying patterns to change in time (e.g., spatial changes in the brain, contextual changes in topics). In this paper, we propose temporal PARAFAC2 (tPARAFAC2): a PARAFAC2-based tensor factorization method with temporal regularization to extract gradually evolving patterns from temporal data. Through extensive experiments on synthetic data, we demonstrate that tPARAFAC2 can capture the underlying evolving patterns accurately performing better than PARAFAC2 and coupled matrix factorization with temporal smoothness regularization.
PARAFAC2-based Coupled Matrix and Tensor Factorizations
Oct 24, 2022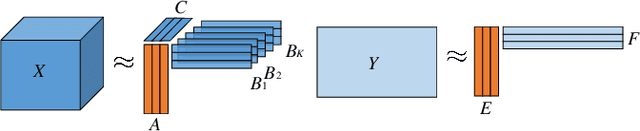
Abstract:Coupled matrix and tensor factorizations (CMTF) have emerged as an effective data fusion tool to jointly analyze data sets in the form of matrices and higher-order tensors. The PARAFAC2 model has shown to be a promising alternative to the CANDECOMP/PARAFAC (CP) tensor model due to its flexibility and capability to handle irregular/ragged tensors. While fusion models based on a PARAFAC2 model coupled with matrix/tensor decompositions have been recently studied, they are limited in terms of possible regularizations and/or types of coupling between data sets. In this paper, we propose an algorithmic framework for fitting PARAFAC2-based CMTF models with the possibility of imposing various constraints on all modes and linear couplings, using Alternating Optimization (AO) and the Alternating Direction Method of Multipliers (ADMM). Through numerical experiments, we demonstrate that the proposed algorithmic approach accurately recovers the underlying patterns using various constraints and linear couplings.
An AO-ADMM approach to constraining PARAFAC2 on all modes
Oct 04, 2021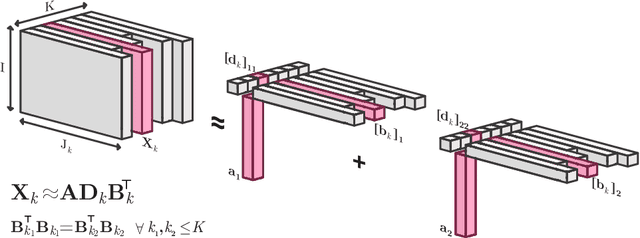
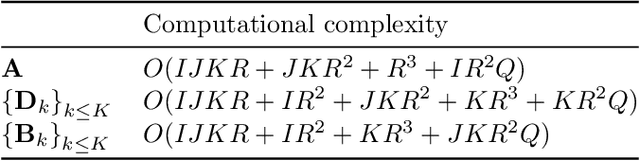
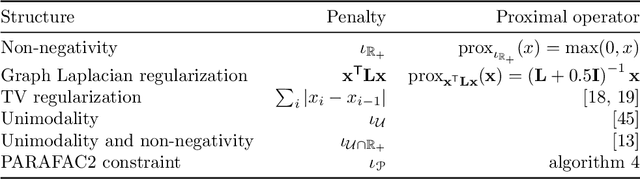
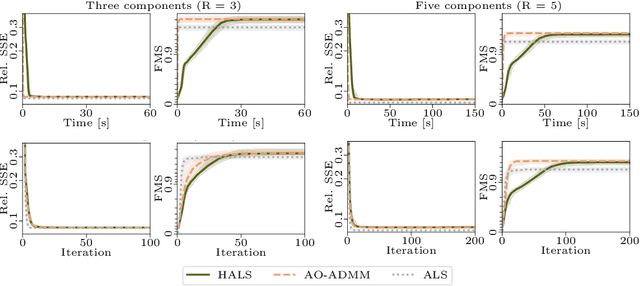
Abstract:Analyzing multi-way measurements with variations across one mode of the dataset is a challenge in various fields including data mining, neuroscience and chemometrics. For example, measurements may evolve over time or have unaligned time profiles. The PARAFAC2 model has been successfully used to analyze such data by allowing the underlying factor matrices in one mode (i.e., the evolving mode) to change across slices. The traditional approach to fit a PARAFAC2 model is to use an alternating least squares-based algorithm, which handles the constant cross-product constraint of the PARAFAC2 model by implicitly estimating the evolving factor matrices. This approach makes imposing regularization on these factor matrices challenging. There is currently no algorithm to flexibly impose such regularization with general penalty functions and hard constraints. In order to address this challenge and to avoid the implicit estimation, in this paper, we propose an algorithm for fitting PARAFAC2 based on alternating optimization with the alternating direction method of multipliers (AO-ADMM). With numerical experiments on simulated data, we show that the proposed PARAFAC2 AO-ADMM approach allows for flexible constraints, recovers the underlying patterns accurately, and is computationally efficient compared to the state-of-the-art. We also apply our model to a real-world chromatography dataset, and show that constraining the evolving mode improves the interpretability of the extracted patterns.
PARAFAC2 AO-ADMM: Constraints in all modes
Feb 03, 2021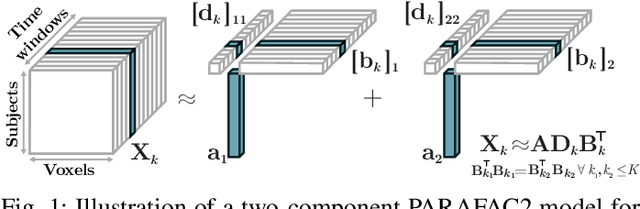
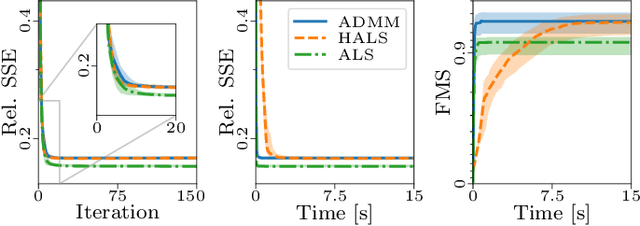
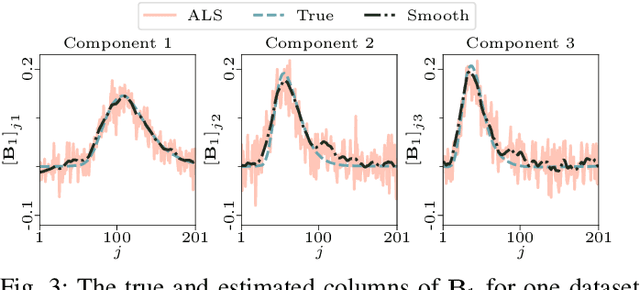
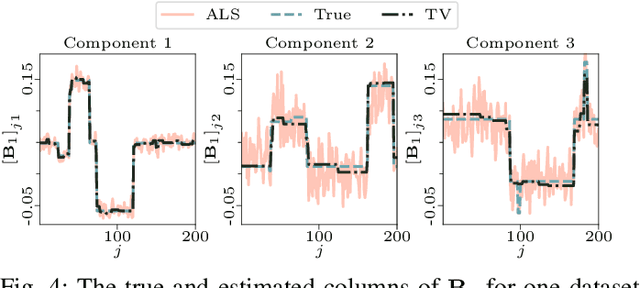
Abstract:The PARAFAC2 model provides a flexible alternative to the popular CANDECOMP/PARAFAC (CP) model for tensor decompositions. Unlike CP, PARAFAC2 allows factor matrices in one mode (i.e., evolving mode) to change across tensor slices, which has proven useful for applications in different domains such as chemometrics, and neuroscience. However, the evolving mode of the PARAFAC2 model is traditionally modelled implicitly, which makes it challenging to regularise it. Currently, the only way to apply regularisation on that mode is with a flexible coupling approach, which finds the solution through regularised least-squares subproblems. In this work, we instead propose an alternating direction method of multipliers (ADMM)-based algorithm for fitting PARAFAC2 and widen the possible regularisation penalties to any proximable function. Our numerical experiments demonstrate that the proposed ADMM-based approach for PARAFAC2 can accurately recover the underlying components from simulated data while being both computationally efficient and flexible in terms of imposing constraints.
A Flexible Optimization Framework for Regularized Matrix-Tensor Factorizations with Linear Couplings
Jul 19, 2020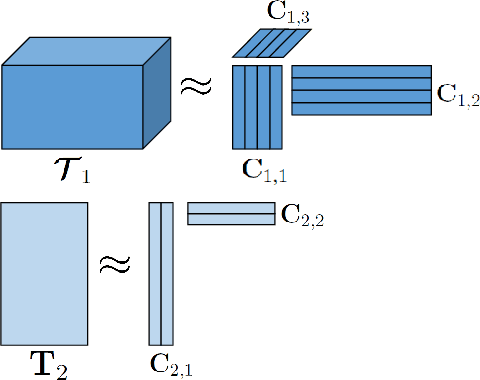
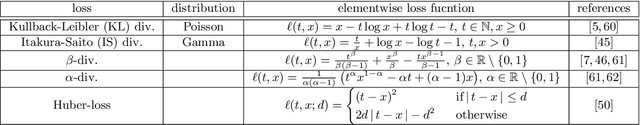


Abstract:Coupled matrix and tensor factorizations (CMTF) are frequently used to jointly analyze data from multiple sources, also called data fusion. However, different characteristics of datasets stemming from multiple sources pose many challenges in data fusion and require to employ various regularizations, constraints, loss functions and different types of coupling structures between datasets. In this paper, we propose a flexible algorithmic framework for coupled matrix and tensor factorizations which utilizes Alternating Optimization (AO) and the Alternating Direction Method of Multipliers (ADMM). The framework facilitates the use of a variety of constraints, loss functions and couplings with linear transformations in a seamless way. Numerical experiments on simulated and real datasets demonstrate that the proposed approach is accurate, and computationally efficient with comparable or better performance than available CMTF methods for Frobenius norm loss, while being more flexible. Using Kullback-Leibler divergence on count data, we demonstrate that the algorithm yields accurate results also for other loss functions.
Tracing Network Evolution Using the PARAFAC2 Model
Oct 23, 2019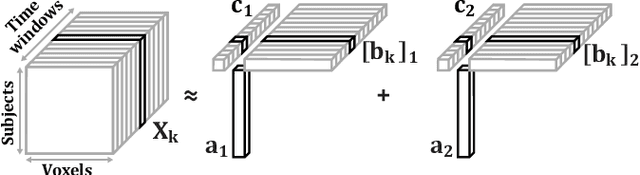
Abstract:Characterizing time-evolving networks is a challenging task, but it is crucial for understanding the dynamic behavior of complex systems such as the brain. For instance, how spatial networks of functional connectivity in the brain evolve during a task is not well-understood. A traditional approach in neuroimaging data analysis is to make simplifications through the assumption of static spatial networks. In this paper, without assuming static networks in time and/or space, we arrange the temporal data as a higher-order tensor and use a tensor factorization model called PARAFAC2 to capture underlying patterns (spatial networks) in time-evolving data and their evolution. Numerical experiments on simulated data demonstrate that PARAFAC2 can successfully reveal the underlying networks and their dynamics. We also show the promising performance of the model in terms of tracing the evolution of task-related functional connectivity in the brain through the analysis of functional magnetic resonance imaging data.
Cross-product Penalized Component Analysis (XCAN)
Jun 28, 2019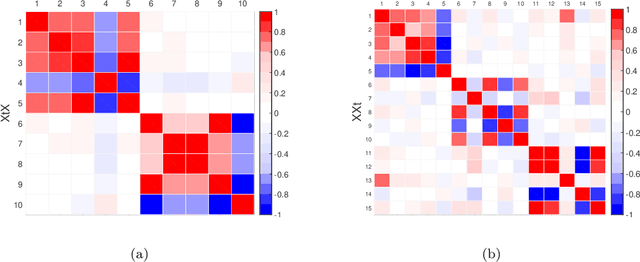
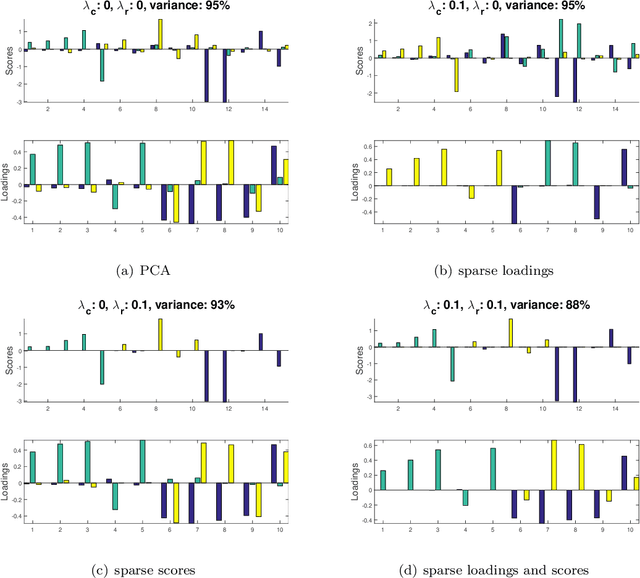
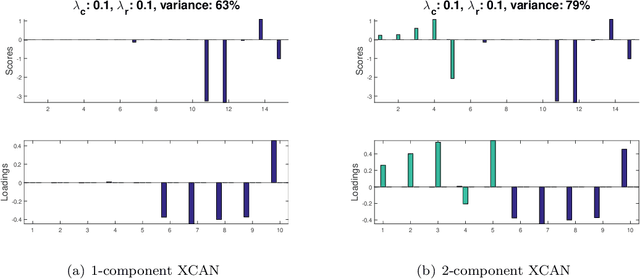
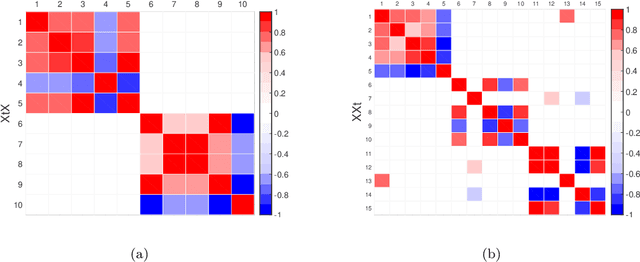
Abstract:Matrix factorization methods are extensively employed to understand complex data. In this paper, we introduce the cross-product penalized component analysis (XCAN), a sparse matrix factorization based on the optimization of a loss function that allows a trade-off between variance maximization and structural preservation. The approach is based on previous developments, notably (i) the Sparse Principal Component Analysis (SPCA) framework based on the LASSO, (ii) extensions of SPCA to constrain both modes of the factorization, like co-clustering or the Penalized Matrix Decomposition (PMD), and (iii) the Group-wise Principal Component Analysis (GPCA) method. The result is a flexible modeling approach that can be used for data exploration in a large variety of problems. We demonstrate its use with applications from different disciplines.
Tensor-Based Fusion of EEG and FMRI to Understand Neurological Changes in Schizophrenia
Dec 07, 2016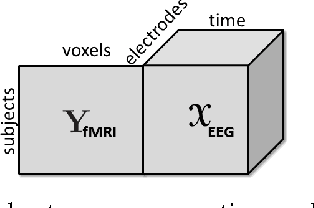
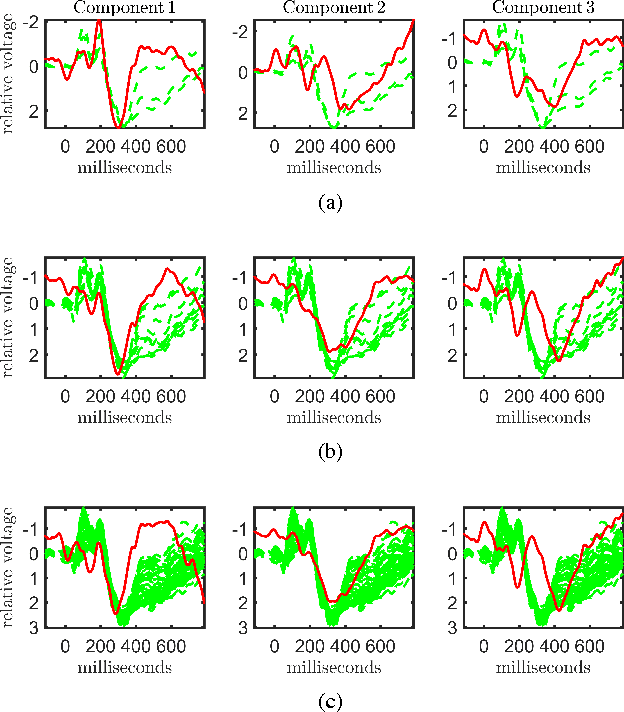
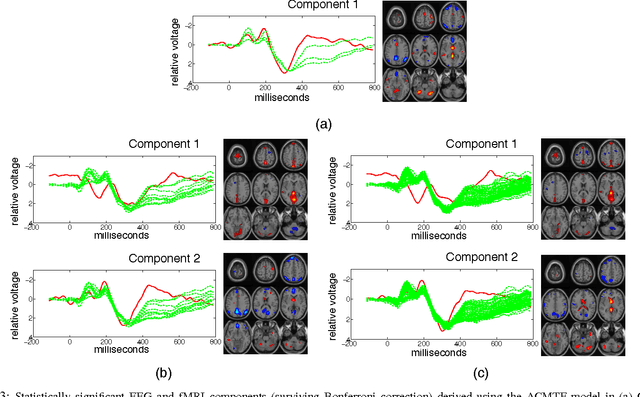
Abstract:Neuroimaging modalities such as functional magnetic resonance imaging (fMRI) and electroencephalography (EEG) provide information about neurological functions in complementary spatiotemporal resolutions; therefore, fusion of these modalities is expected to provide better understanding of brain activity. In this paper, we jointly analyze fMRI and multi-channel EEG signals collected during an auditory oddball task with the goal of capturing brain activity patterns that differ between patients with schizophrenia and healthy controls. Rather than selecting a single electrode or matricizing the third-order tensor that can be naturally used to represent multi-channel EEG signals, we preserve the multi-way structure of EEG data and use a coupled matrix and tensor factorization (CMTF) model to jointly analyze fMRI and EEG signals. Our analysis reveals that (i) joint analysis of EEG and fMRI using a CMTF model can capture meaningful temporal and spatial signatures of patterns that behave differently in patients and controls, and (ii) these differences and the interpretability of the associated components increase by including multiple electrodes from frontal, motor and parietal areas, but not necessarily by including all electrodes in the analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge