Marie Roald
Comparative analysis of optical character recognition methods for Sámi texts from the National Library of Norway
Jan 13, 2025



Abstract:Optical Character Recognition (OCR) is crucial to the National Library of Norway's (NLN) digitisation process as it converts scanned documents into machine-readable text. However, for the S\'ami documents in NLN's collection, the OCR accuracy is insufficient. Given that OCR quality affects downstream processes, evaluating and improving OCR for text written in S\'ami languages is necessary to make these resources accessible. To address this need, this work fine-tunes and evaluates three established OCR approaches, Transkribus, Tesseract and TrOCR, for transcribing S\'ami texts from NLN's collection. Our results show that Transkribus and TrOCR outperform Tesseract on this task, while Tesseract achieves superior performance on an out-of-domain dataset. Furthermore, we show that fine-tuning pre-trained models and supplementing manual annotations with machine annotations and synthetic text images can yield accurate OCR for S\'ami languages, even with a moderate amount of manually annotated data.
Visual Navigation of Digital Libraries: Retrieval and Classification of Images in the National Library of Norway's Digitised Book Collection
Oct 19, 2024



Abstract:Digital tools for text analysis have long been essential for the searchability and accessibility of digitised library collections. Recent computer vision advances have introduced similar capabilities for visual materials, with deep learning-based embeddings showing promise for analysing visual heritage. Given that many books feature visuals in addition to text, taking advantage of these breakthroughs is critical to making library collections open and accessible. In this work, we present a proof-of-concept image search application for exploring images in the National Library of Norway's pre-1900 books, comparing Vision Transformer (ViT), Contrastive Language-Image Pre-training (CLIP), and Sigmoid loss for Language-Image Pre-training (SigLIP) embeddings for image retrieval and classification. Our results show that the application performs well for exact image retrieval, with SigLIP embeddings slightly outperforming CLIP and ViT in both retrieval and classification tasks. Additionally, SigLIP-based image classification can aid in cleaning image datasets from a digitisation pipeline.
An AO-ADMM approach to constraining PARAFAC2 on all modes
Oct 04, 2021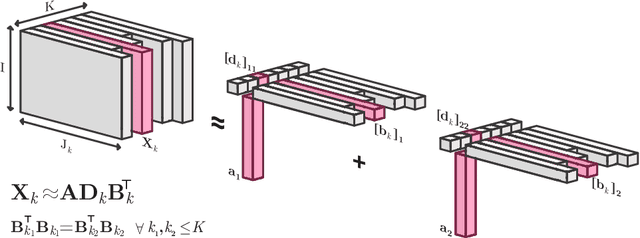
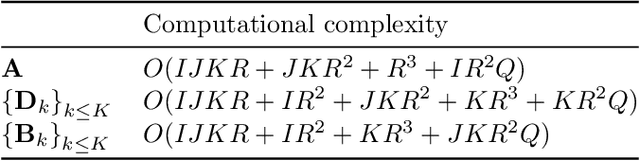
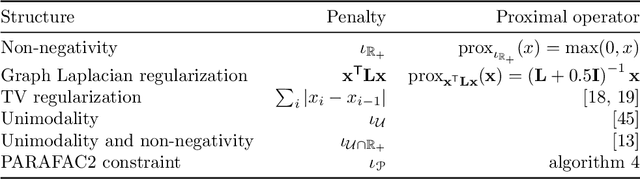
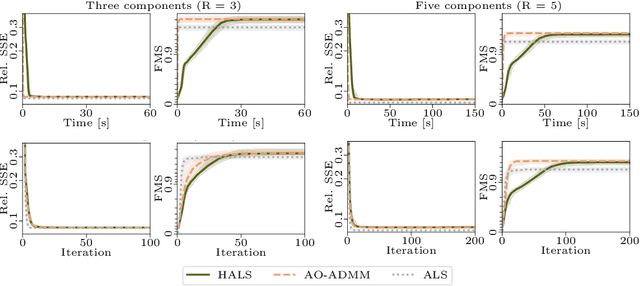
Abstract:Analyzing multi-way measurements with variations across one mode of the dataset is a challenge in various fields including data mining, neuroscience and chemometrics. For example, measurements may evolve over time or have unaligned time profiles. The PARAFAC2 model has been successfully used to analyze such data by allowing the underlying factor matrices in one mode (i.e., the evolving mode) to change across slices. The traditional approach to fit a PARAFAC2 model is to use an alternating least squares-based algorithm, which handles the constant cross-product constraint of the PARAFAC2 model by implicitly estimating the evolving factor matrices. This approach makes imposing regularization on these factor matrices challenging. There is currently no algorithm to flexibly impose such regularization with general penalty functions and hard constraints. In order to address this challenge and to avoid the implicit estimation, in this paper, we propose an algorithm for fitting PARAFAC2 based on alternating optimization with the alternating direction method of multipliers (AO-ADMM). With numerical experiments on simulated data, we show that the proposed PARAFAC2 AO-ADMM approach allows for flexible constraints, recovers the underlying patterns accurately, and is computationally efficient compared to the state-of-the-art. We also apply our model to a real-world chromatography dataset, and show that constraining the evolving mode improves the interpretability of the extracted patterns.
PARAFAC2 AO-ADMM: Constraints in all modes
Feb 03, 2021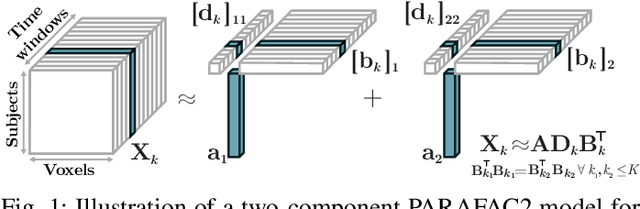
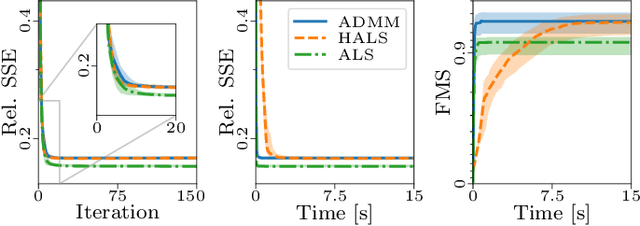
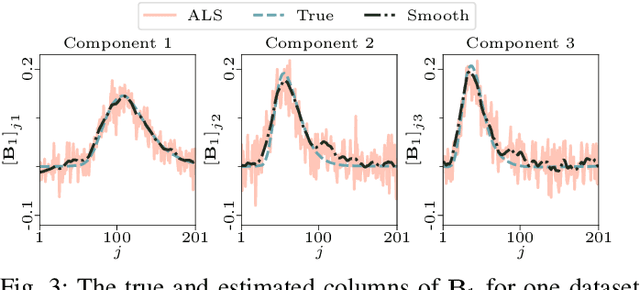
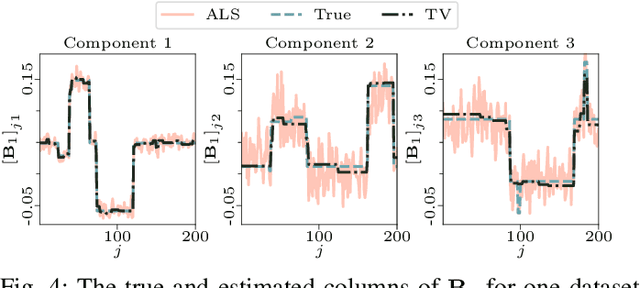
Abstract:The PARAFAC2 model provides a flexible alternative to the popular CANDECOMP/PARAFAC (CP) model for tensor decompositions. Unlike CP, PARAFAC2 allows factor matrices in one mode (i.e., evolving mode) to change across tensor slices, which has proven useful for applications in different domains such as chemometrics, and neuroscience. However, the evolving mode of the PARAFAC2 model is traditionally modelled implicitly, which makes it challenging to regularise it. Currently, the only way to apply regularisation on that mode is with a flexible coupling approach, which finds the solution through regularised least-squares subproblems. In this work, we instead propose an alternating direction method of multipliers (ADMM)-based algorithm for fitting PARAFAC2 and widen the possible regularisation penalties to any proximable function. Our numerical experiments demonstrate that the proposed ADMM-based approach for PARAFAC2 can accurately recover the underlying components from simulated data while being both computationally efficient and flexible in terms of imposing constraints.
Tracing Network Evolution Using the PARAFAC2 Model
Oct 23, 2019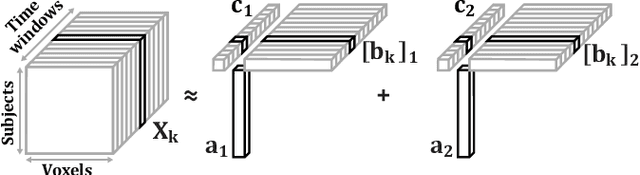
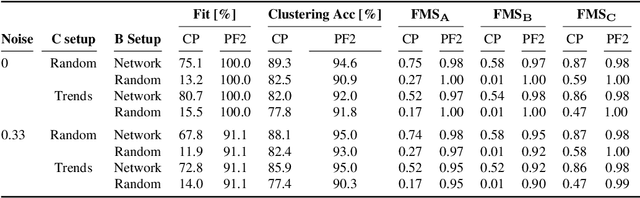
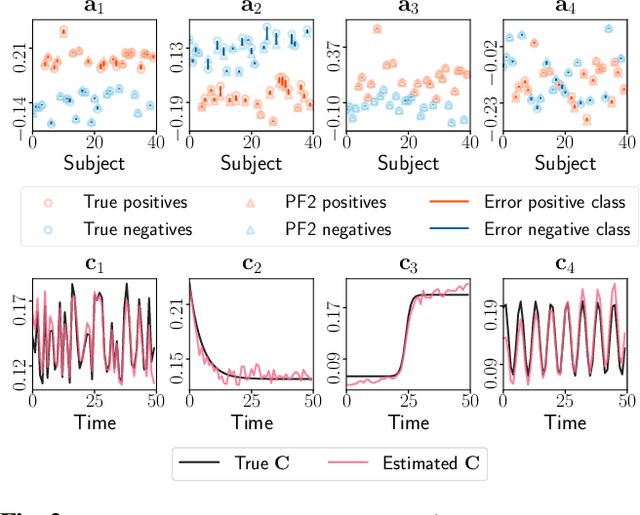
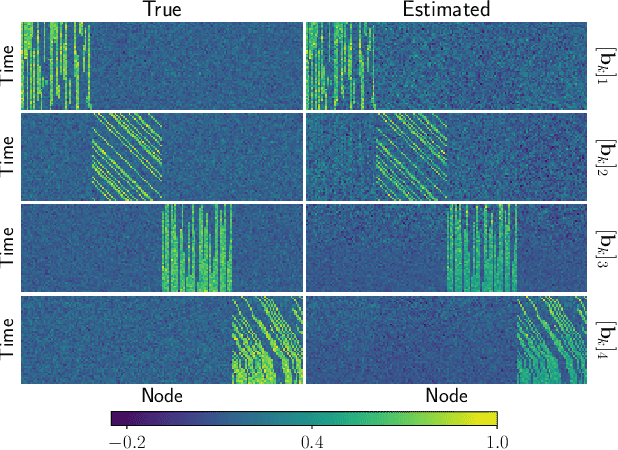
Abstract:Characterizing time-evolving networks is a challenging task, but it is crucial for understanding the dynamic behavior of complex systems such as the brain. For instance, how spatial networks of functional connectivity in the brain evolve during a task is not well-understood. A traditional approach in neuroimaging data analysis is to make simplifications through the assumption of static spatial networks. In this paper, without assuming static networks in time and/or space, we arrange the temporal data as a higher-order tensor and use a tensor factorization model called PARAFAC2 to capture underlying patterns (spatial networks) in time-evolving data and their evolution. Numerical experiments on simulated data demonstrate that PARAFAC2 can successfully reveal the underlying networks and their dynamics. We also show the promising performance of the model in terms of tracing the evolution of task-related functional connectivity in the brain through the analysis of functional magnetic resonance imaging data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge