Donald Brown
The American Ghost in the Machine: How language models align culturally and the effects of cultural prompting
Dec 13, 2025Abstract:Culture is the bedrock of human interaction; it dictates how we perceive and respond to everyday interactions. As the field of human-computer interaction grows via the rise of generative Large Language Models (LLMs), the cultural alignment of these models become an important field of study. This work, using the VSM13 International Survey and Hofstede's cultural dimensions, identifies the cultural alignment of popular LLMs (DeepSeek-V3, V3.1, GPT-5, GPT-4.1, GPT-4, Claude Opus 4, Llama 3.1, and Mistral Large). We then use cultural prompting, or using system prompts to shift the cultural alignment of a model to a desired country, to test the adaptability of these models to other cultures, namely China, France, India, Iran, Japan, and the United States. We find that the majority of the eight LLMs tested favor the United States when the culture is not specified, with varying results when prompted for other cultures. When using cultural prompting, seven of the eight models shifted closer to the expected culture. We find that models had trouble aligning with Japan and China, despite two of the models tested originating with the Chinese company DeepSeek.
DeepSeek's WEIRD Behavior: The cultural alignment of Large Language Models and the effects of prompt language and cultural prompting
Dec 12, 2025



Abstract:Culture is a core component of human-to-human interaction and plays a vital role in how we perceive and interact with others. Advancements in the effectiveness of Large Language Models (LLMs) in generating human-sounding text have greatly increased the amount of human-to-computer interaction. As this field grows, the cultural alignment of these human-like agents becomes an important field of study. Our work uses Hofstede's VSM13 international surveys to understand the cultural alignment of the following models: DeepSeek-V3, V3.1, GPT-4, GPT-4.1, GPT-4o, and GPT-5. We use a combination of prompt language and cultural prompting, a strategy that uses a system prompt to shift a model's alignment to reflect a specific country, to align these LLMs with the United States and China. Our results show that DeepSeek-V3, V3.1, and OpenAI's GPT-5 exhibit a close alignment with the survey responses of the United States and do not achieve a strong or soft alignment with China, even when using cultural prompts or changing the prompt language. We also find that GPT-4 exhibits an alignment closer to China when prompted in English, but cultural prompting is effective in shifting this alignment closer to the United States. Other low-cost models, GPT-4o and GPT-4.1, respond to the prompt language used (i.e., English or Simplified Chinese) and cultural prompting strategies to create acceptable alignments with both the United States and China.
Diffusion and Multi-Domain Adaptation Methods for Eosinophil Segmentation
Mar 17, 2024Abstract:Eosinophilic Esophagitis (EoE) represents a challenging condition for medical providers today. The cause is currently unknown, the impact on a patient's daily life is significant, and it is increasing in prevalence. Traditional approaches for medical image diagnosis such as standard deep learning algorithms are limited by the relatively small amount of data and difficulty in generalization. As a response, two methods have arisen that seem to perform well: Diffusion and Multi-Domain methods with current research efforts favoring diffusion methods. For the EoE dataset, we discovered that a Multi-Domain Adversarial Network outperformed a Diffusion based method with a FID of 42.56 compared to 50.65. Future work with diffusion methods should include a comparison with Multi-Domain adaptation methods to ensure that the best performance is achieved.
Uncertainty Quantification for Eosinophil Segmentation
Sep 28, 2023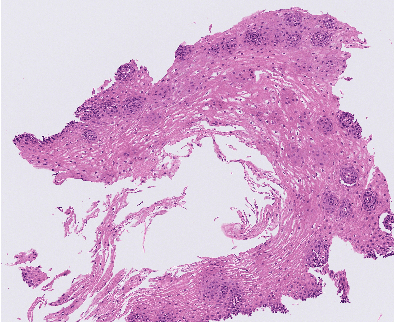
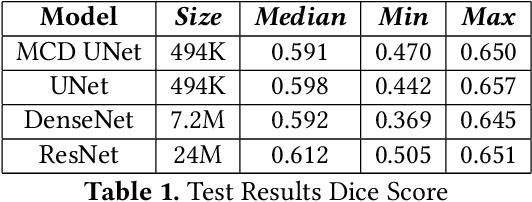
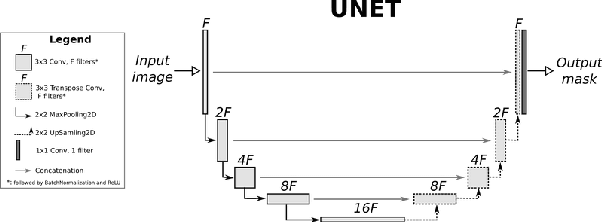
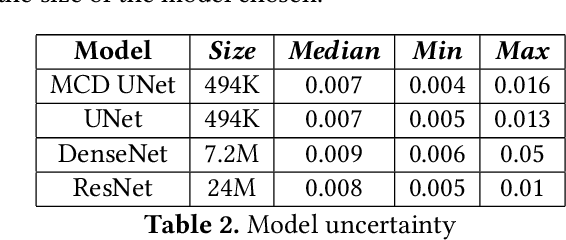
Abstract:Eosinophilic Esophagitis (EoE) is an allergic condition increasing in prevalence. To diagnose EoE, pathologists must find 15 or more eosinophils within a single high-power field (400X magnification). Determining whether or not a patient has EoE can be an arduous process and any medical imaging approaches used to assist diagnosis must consider both efficiency and precision. We propose an improvement of Adorno et al's approach for quantifying eosinphils using deep image segmentation. Our new approach leverages Monte Carlo Dropout, a common approach in deep learning to reduce overfitting, to provide uncertainty quantification on current deep learning models. The uncertainty can be visualized in an output image to evaluate model performance, provide insight to how deep learning algorithms function, and assist pathologists in identifying eosinophils.
The intersection of video capsule endoscopy and artificial intelligence: addressing unique challenges using machine learning
Aug 24, 2023



Abstract:Introduction: Technical burdens and time-intensive review processes limit the practical utility of video capsule endoscopy (VCE). Artificial intelligence (AI) is poised to address these limitations, but the intersection of AI and VCE reveals challenges that must first be overcome. We identified five challenges to address. Challenge #1: VCE data are stochastic and contains significant artifact. Challenge #2: VCE interpretation is cost-intensive. Challenge #3: VCE data are inherently imbalanced. Challenge #4: Existing VCE AIMLT are computationally cumbersome. Challenge #5: Clinicians are hesitant to accept AIMLT that cannot explain their process. Methods: An anatomic landmark detection model was used to test the application of convolutional neural networks (CNNs) to the task of classifying VCE data. We also created a tool that assists in expert annotation of VCE data. We then created more elaborate models using different approaches including a multi-frame approach, a CNN based on graph representation, and a few-shot approach based on meta-learning. Results: When used on full-length VCE footage, CNNs accurately identified anatomic landmarks (99.1%), with gradient weighted-class activation mapping showing the parts of each frame that the CNN used to make its decision. The graph CNN with weakly supervised learning (accuracy 89.9%, sensitivity of 91.1%), the few-shot model (accuracy 90.8%, precision 91.4%, sensitivity 90.9%), and the multi-frame model (accuracy 97.5%, precision 91.5%, sensitivity 94.8%) performed well. Discussion: Each of these five challenges is addressed, in part, by one of our AI-based models. Our goal of producing high performance using lightweight models that aim to improve clinician confidence was achieved.
Graph Convolution Neural Network For Weakly Supervised Abnormality Localization In Long Capsule Endoscopy Videos
Oct 18, 2021
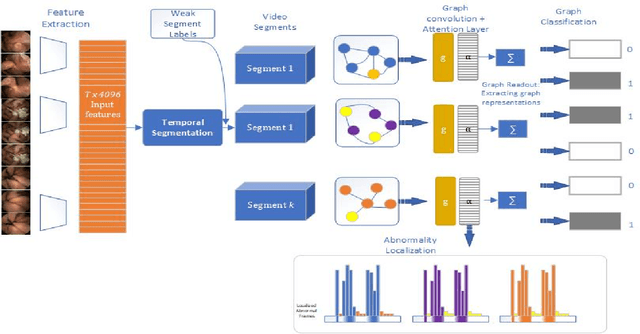
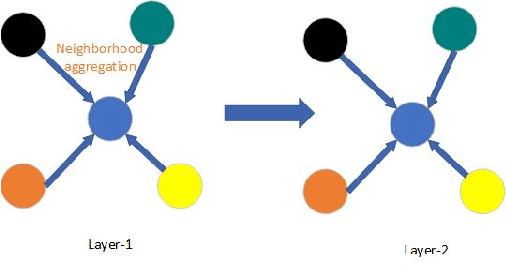
Abstract:Temporal activity localization in long videos is an important problem. The cost of obtaining frame level label for long Wireless Capsule Endoscopy (WCE) videos is prohibitive. In this paper, we propose an end-to-end temporal abnormality localization for long WCE videos using only weak video level labels. Physicians use Capsule Endoscopy (CE) as a non-surgical and non-invasive method to examine the entire digestive tract in order to diagnose diseases or abnormalities. While CE has revolutionized traditional endoscopy procedures, a single CE examination could last up to 8 hours generating as much as 100,000 frames. Physicians must review the entire video, frame-by-frame, in order to identify the frames capturing relevant abnormality. This, sometimes could be as few as just a single frame. Given this very high level of redundancy, analyzing long CE videos can be very tedious, time consuming and also error prone. This paper presents a novel multi-step method for an end-to-end localization of target frames capturing abnormalities of interest in the long video using only weak video labels. First we developed an automatic temporal segmentation using change point detection technique to temporally segment the video into uniform, homogeneous and identifiable segments. Then we employed Graph Convolutional Neural Network (GCNN) to learn a representation of each video segment. Using weak video segment labels, we trained our GCNN model to recognize each video segment as abnormal if it contains at least a single abnormal frame. Finally, leveraging the parameters of the trained GCNN model, we replaced the final layer of the network with a temporal pool layer to localize the relevant abnormal frames within each abnormal video segment. Our method achieved an accuracy of 89.9\% on the graph classification task and a specificity of 97.5\% on the abnormal frames localization task.
Unsupervised Shot Boundary Detection for Temporal Segmentation of Long Capsule Endoscopy Videos
Oct 18, 2021



Abstract:Physicians use Capsule Endoscopy (CE) as a non-invasive and non-surgical procedure to examine the entire gastrointestinal (GI) tract for diseases and abnormalities. A single CE examination could last between 8 to 11 hours generating up to 80,000 frames which is compiled as a video. Physicians have to review and analyze the entire video to identify abnormalities or diseases before making diagnosis. This review task can be very tedious, time consuming and prone to error. While only as little as a single frame may capture useful content that is relevant to the physicians' final diagnosis, frames covering the small bowel region alone could be as much as 50,000. To minimize physicians' review time and effort, this paper proposes a novel unsupervised and computationally efficient temporal segmentation method to automatically partition long CE videos into a homogeneous and identifiable video segments. However, the search for temporal boundaries in a long video using high dimensional frame-feature matrix is computationally prohibitive and impracticable for real clinical application. Therefore, leveraging both spatial and temporal information in the video, we first extracted high level frame features using a pretrained CNN model and then projected the high-dimensional frame-feature matrix to lower 1-dimensional embedding. Using this 1-dimensional sequence embedding, we applied the Pruned Exact Linear Time (PELT) algorithm to searched for temporal boundaries that indicates the transition points from normal to abnormal frames and vice-versa. We experimented with multiple real patients' CE videos and our model achieved an AUC of 66\% on multiple test videos against expert provided labels.
Lesion2Vec: Deep Metric Learning for Few-Shot Multiple Lesions Recognition in Wireless Capsule Endoscopy Video
Jan 15, 2021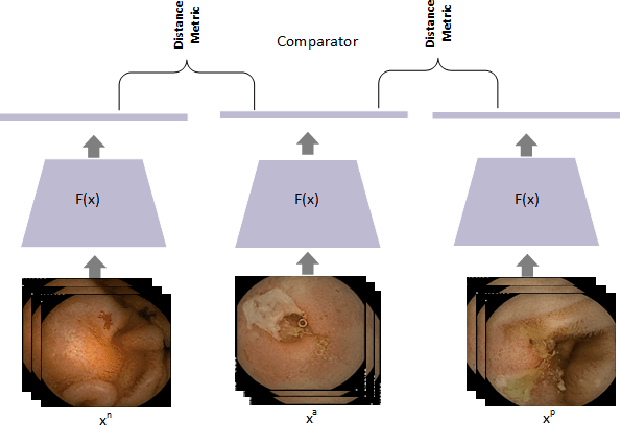
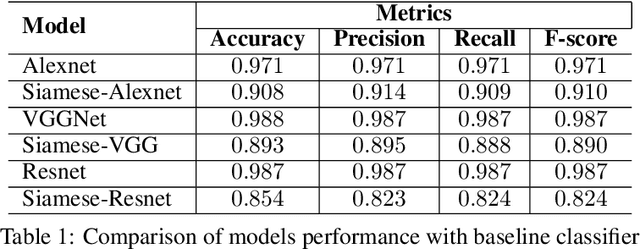
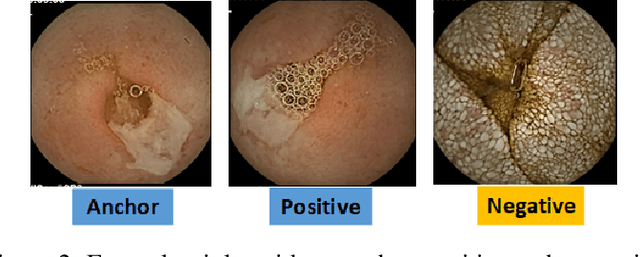
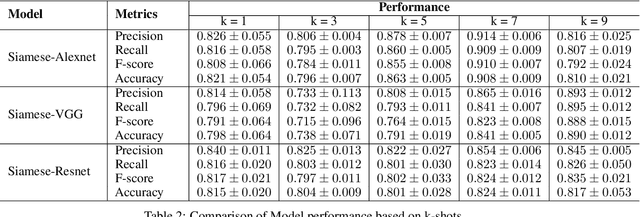
Abstract:Effective and rapid detection of lesions in the Gastrointestinal tract is critical to gastroenterologist's response to some life-threatening diseases. Wireless Capsule Endoscopy (WCE) has revolutionized traditional endoscopy procedure by allowing gastroenterologists visualize the entire GI tract non-invasively. Once the tiny capsule is swallowed, it sequentially capture images of the GI tract at about 2 to 6 frames per second (fps). A single video can last up to 8 hours producing between 30,000 to 100,000 images. Automating the detection of frames containing specific lesion in WCE video would relieve gastroenterologists the arduous task of reviewing the entire video before making diagnosis. While the WCE produces large volume of images, only about 5\% of the frames contain lesions that aid the diagnosis process. Convolutional Neural Network (CNN) based models have been very successful in various image classification tasks. However, they suffer excessive parameters, are sample inefficient and rely on very large amount of training data. Deploying a CNN classifier for lesion detection task will require time-to-time fine-tuning to generalize to any unforeseen category. In this paper, we propose a metric-based learning framework followed by a few-shot lesion recognition in WCE data. Metric-based learning is a meta-learning framework designed to establish similarity or dissimilarity between concepts while few-shot learning (FSL) aims to identify new concepts from only a small number of examples. We train a feature extractor to learn a representation for different small bowel lesions using metric-based learning. At the testing stage, the category of an unseen sample is predicted from only a few support examples, thereby allowing the model to generalize to a new category that has never been seen before. We demonstrated the efficacy of this method on real patient capsule endoscopy data.
Improving Classification through Weak Supervision in Context-specific Conversational Agent Development for Teacher Education
Oct 23, 2020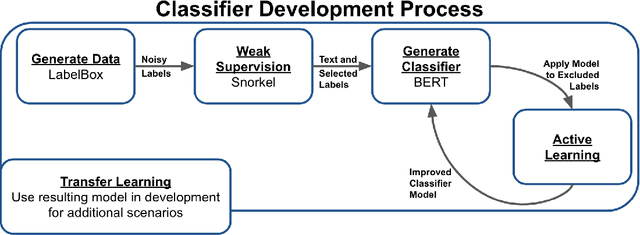

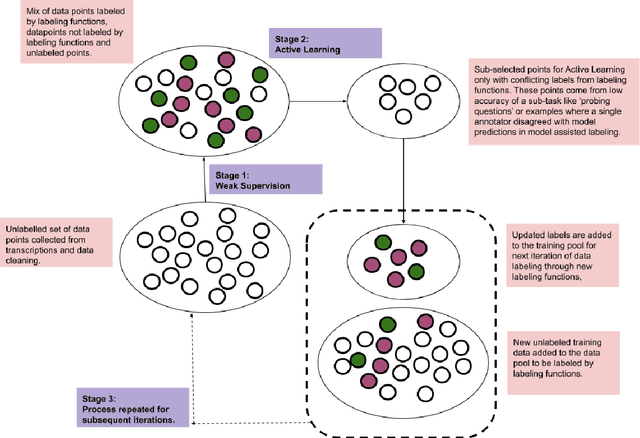

Abstract:Machine learning techniques applied to the Natural Language Processing (NLP) component of conversational agent development show promising results for improved accuracy and quality of feedback that a conversational agent can provide. The effort required to develop an educational scenario specific conversational agent is time consuming as it requires domain experts to label and annotate noisy data sources such as classroom videos. Previous approaches to modeling annotations have relied on labeling thousands of examples and calculating inter-annotator agreement and majority votes in order to model the necessary scenarios. This method, while proven successful, ignores individual annotator strengths in labeling a data point and under-utilizes examples that do not have a majority vote for labeling. We propose using a multi-task weak supervision method combined with active learning to address these concerns. This approach requires less labeling than traditional methods and shows significant improvements in precision, efficiency, and time-requirements than the majority vote method (Ratner 2019). We demonstrate the validity of this method on the Google Jigsaw data set and then propose a scenario to apply this method using the Instructional Quality Assessment(IQA) to define the categories for labeling. We propose using probabilistic modeling of annotator labeling to generate active learning examples to further label the data. Active learning is able to iteratively improve the training performance and accuracy of the original classification model. This approach combines state-of-the art labeling techniques of weak supervision and active learning to optimize results in the educational domain and could be further used to lessen the data requirements for expanded scenarios within the education domain through transfer learning.
Hand-drawn Symbol Recognition of Surgical Flowsheet Graphs with Deep Image Segmentation
Jun 30, 2020

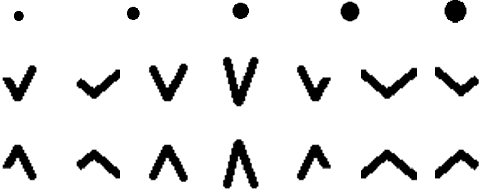
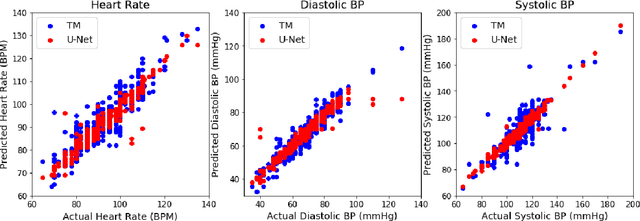
Abstract:Perioperative data are essential to investigating the causes of adverse surgical outcomes. In some low to middle income countries, these data are computationally inaccessible due to a lack of digitization of surgical flowsheets. In this paper, we present a deep image segmentation approach using a U-Net architecture that can detect hand-drawn symbols on a flowsheet graph. The segmentation mask outputs are post-processed with techniques unique to each symbol to convert into numeric values. The U-Net method can detect, at the appropriate time intervals, the symbols for heart rate and blood pressure with over 99 percent accuracy. Over 95 percent of the predictions fall within an absolute error of five when compared to the actual value. The deep learning model outperformed template matching even with a small size of annotated images available for the training set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge