Chun-Hung Chao
Self-supervised Landmark Learning with Deformation Reconstruction and Cross-subject Consistency Objectives
Aug 09, 2023Abstract:A Point Distribution Model (PDM) is the basis of a Statistical Shape Model (SSM) that relies on a set of landmark points to represent a shape and characterize the shape variation. In this work, we present a self-supervised approach to extract landmark points from a given registration model for the PDMs. Based on the assumption that the landmarks are the points that have the most influence on registration, existing works learn a point-based registration model with a small number of points to estimate the landmark points that influence the deformation the most. However, such approaches assume that the deformation can be captured by point-based registration and quality landmarks can be learned solely with the deformation capturing objective. We argue that data with complicated deformations can not easily be modeled with point-based registration when only a limited number of points is used to extract influential landmark points. Further, landmark consistency is not assured in existing approaches In contrast, we propose to extract landmarks based on a given registration model, which is tailored for the target data, so we can obtain more accurate correspondences. Secondly, to establish the anatomical consistency of the predicted landmarks, we introduce a landmark discovery loss to explicitly encourage the model to predict the landmarks that are anatomically consistent across subjects. We conduct experiments on an osteoarthritis progression prediction task and show our method outperforms existing image-based and point-based approaches.
Interactive Radiotherapy Target Delineation with 3D-Fused Context Propagation
Dec 12, 2020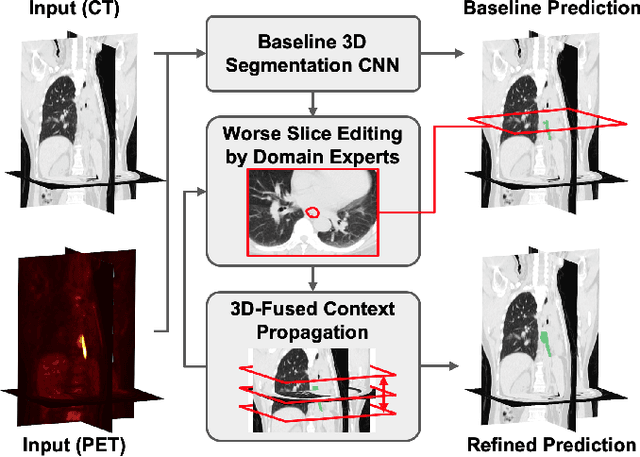
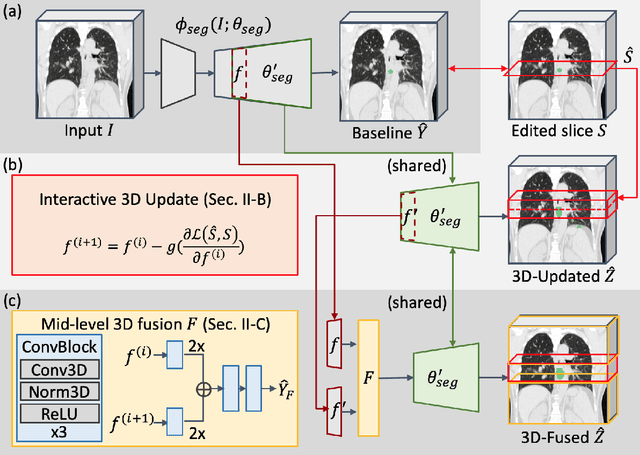

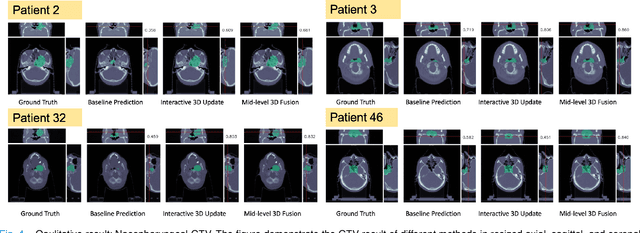
Abstract:Gross tumor volume (GTV) delineation on tomography medical imaging is crucial for radiotherapy planning and cancer diagnosis. Convolutional neural networks (CNNs) has been predominated on automatic 3D medical segmentation tasks, including contouring the radiotherapy target given 3D CT volume. While CNNs may provide feasible outcome, in clinical scenario, double-check and prediction refinement by experts is still necessary because of CNNs' inconsistent performance on unexpected patient cases. To provide experts an efficient way to modify the CNN predictions without retrain the model, we propose 3D-fused context propagation, which propagates any edited slice to the whole 3D volume. By considering the high-level feature maps, the radiation oncologists would only required to edit few slices to guide the correction and refine the whole prediction volume. Specifically, we leverage the backpropagation for activation technique to convey the user editing information backwardly to the latent space and generate new prediction based on the updated and original feature. During the interaction, our proposed approach reuses the extant extracted features and does not alter the existing 3D CNN model architectures, avoiding the perturbation on other predictions. The proposed method is evaluated on two published radiotherapy target contouring datasets of nasopharyngeal and esophageal cancer. The experimental results demonstrate that our proposed method is able to further effectively improve the existing segmentation prediction from different model architectures given oncologists' interactive inputs.
Lymph Node Gross Tumor Volume Detection in Oncology Imaging via Relationship Learning Using Graph Neural Network
Aug 29, 2020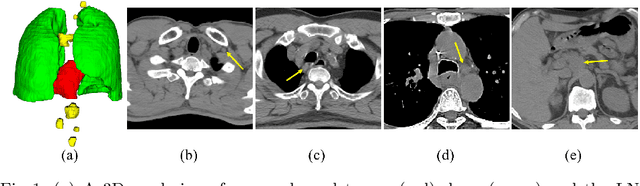
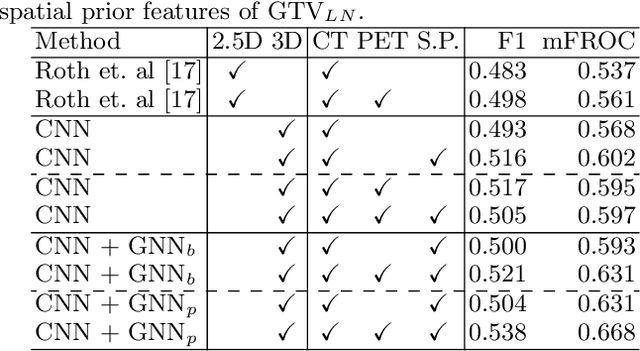
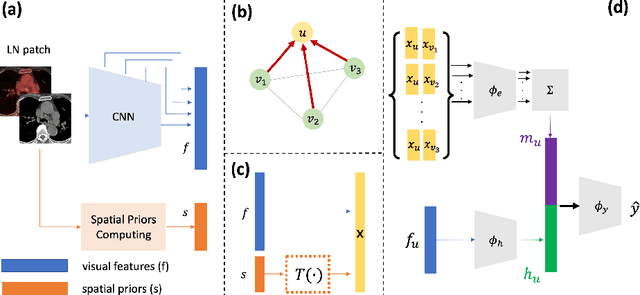
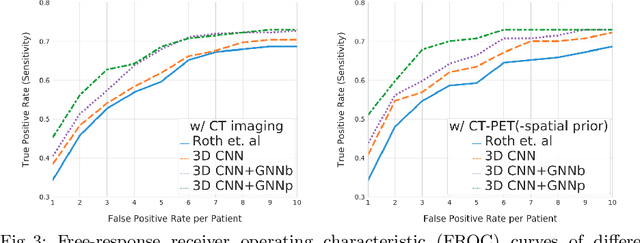
Abstract:Determining the spread of GTV$_{LN}$ is essential in defining the respective resection or irradiating regions for the downstream workflows of surgical resection and radiotherapy for many cancers. Different from the more common enlarged lymph node (LN), GTV$_{LN}$ also includes smaller ones if associated with high positron emission tomography signals and/or any metastasis signs in CT. This is a daunting task. In this work, we propose a unified LN appearance and inter-LN relationship learning framework to detect the true GTV$_{LN}$. This is motivated by the prior clinical knowledge that LNs form a connected lymphatic system, and the spread of cancer cells among LNs often follows certain pathways. Specifically, we first utilize a 3D convolutional neural network with ROI-pooling to extract the GTV$_{LN}$'s instance-wise appearance features. Next, we introduce a graph neural network to further model the inter-LN relationships where the global LN-tumor spatial priors are included in the learning process. This leads to an end-to-end trainable network to detect by classifying GTV$_{LN}$. We operate our model on a set of GTV$_{LN}$ candidates generated by a preliminary 1st-stage method, which has a sensitivity of $>85\%$ at the cost of high false positive (FP) ($>15$ FPs per patient). We validate our approach on a radiotherapy dataset with 142 paired PET/RTCT scans containing the chest and upper abdominal body parts. The proposed method significantly improves over the state-of-the-art (SOTA) LN classification method by $5.5\%$ and $13.1\%$ in F1 score and the averaged sensitivity value at $2, 3, 4, 6$ FPs per patient, respectively.
Lymph Node Gross Tumor Volume Detection and Segmentation via Distance-based Gating using 3D CT/PET Imaging in Radiotherapy
Aug 27, 2020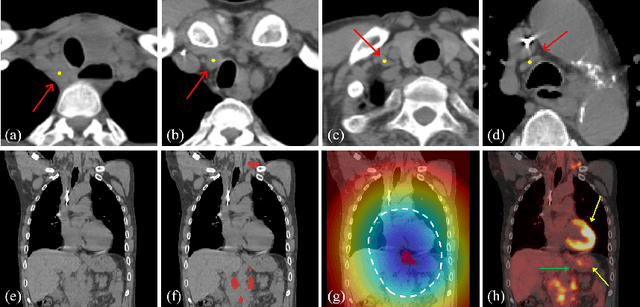
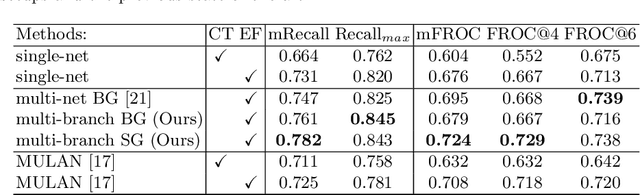
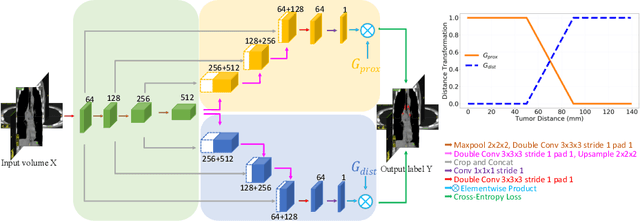
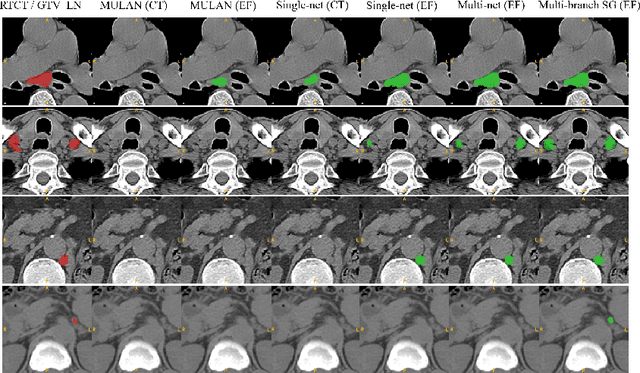
Abstract:Finding, identifying and segmenting suspicious cancer metastasized lymph nodes from 3D multi-modality imaging is a clinical task of paramount importance. In radiotherapy, they are referred to as Lymph Node Gross Tumor Volume (GTVLN). Determining and delineating the spread of GTVLN is essential in defining the corresponding resection and irradiating regions for the downstream workflows of surgical resection and radiotherapy of various cancers. In this work, we propose an effective distance-based gating approach to simulate and simplify the high-level reasoning protocols conducted by radiation oncologists, in a divide-and-conquer manner. GTVLN is divided into two subgroups of tumor-proximal and tumor-distal, respectively, by means of binary or soft distance gating. This is motivated by the observation that each category can have distinct though overlapping distributions of appearance, size and other LN characteristics. A novel multi-branch detection-by-segmentation network is trained with each branch specializing on learning one GTVLN category features, and outputs from multi-branch are fused in inference. The proposed method is evaluated on an in-house dataset of $141$ esophageal cancer patients with both PET and CT imaging modalities. Our results validate significant improvements on the mean recall from $72.5\%$ to $78.2\%$, as compared to previous state-of-the-art work. The highest achieved GTVLN recall of $82.5\%$ at $20\%$ precision is clinically relevant and valuable since human observers tend to have low sensitivity (around $80\%$ for the most experienced radiation oncologists, as reported by literature).
Detecting Scatteredly-Distributed, Small, andCritically Important Objects in 3D OncologyImaging via Decision Stratification
May 27, 2020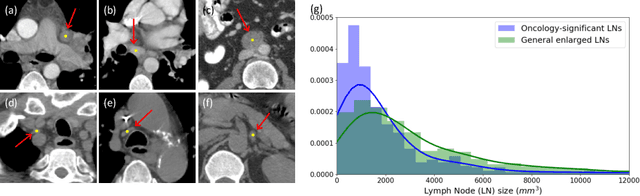
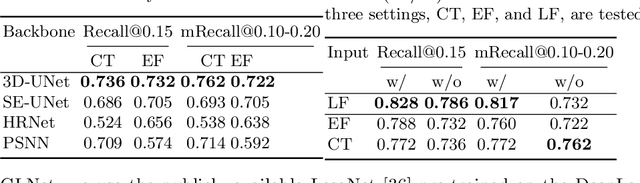
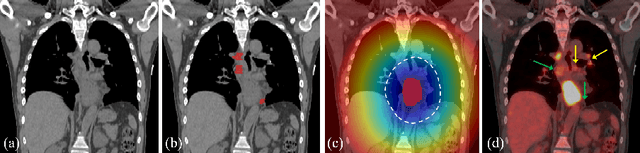
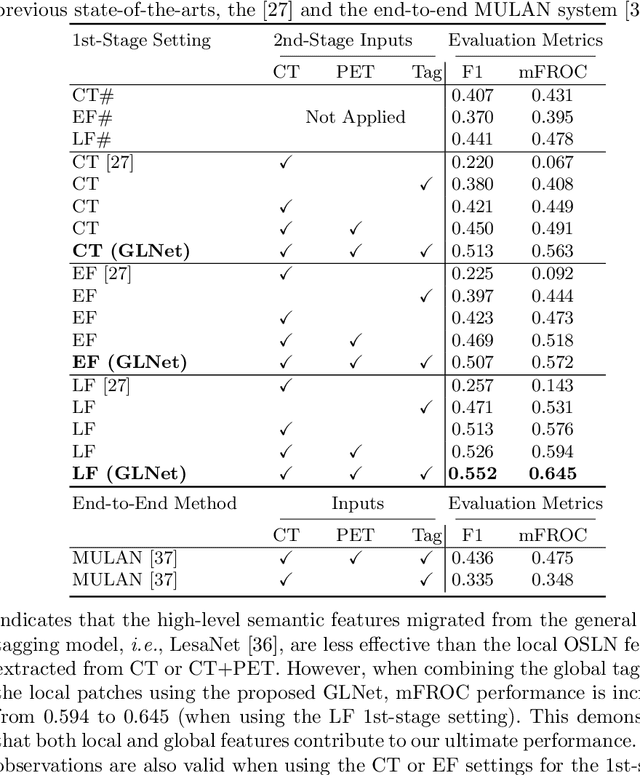
Abstract:Finding and identifying scatteredly-distributed, small, and critically important objects in 3D oncology images is very challenging. We focus on the detection and segmentation of oncology-significant (or suspicious cancer metastasized) lymph nodes (OSLNs), which has not been studied before as a computational task. Determining and delineating the spread of OSLNs is essential in defining the corresponding resection/irradiating regions for the downstream workflows of surgical resection and radiotherapy of various cancers. For patients who are treated with radiotherapy, this task is performed by experienced radiation oncologists that involves high-level reasoning on whether LNs are metastasized, which is subject to high inter-observer variations. In this work, we propose a divide-and-conquer decision stratification approach that divides OSLNs into tumor-proximal and tumor-distal categories. This is motivated by the observation that each category has its own different underlying distributions in appearance, size and other characteristics. Two separate detection-by-segmentation networks are trained per category and fused. To further reduce false positives (FP), we present a novel global-local network (GLNet) that combines high-level lesion characteristics with features learned from localized 3D image patches. Our method is evaluated on a dataset of 141 esophageal cancer patients with PET and CT modalities (the largest to-date). Our results significantly improve the recall from $45\%$ to $67\%$ at $3$ FPs per patient as compared to previous state-of-the-art methods. The highest achieved OSLN recall of $0.828$ is clinically relevant and valuable.
Organ at Risk Segmentation for Head and Neck Cancer using Stratified Learning and Neural Architecture Search
Apr 17, 2020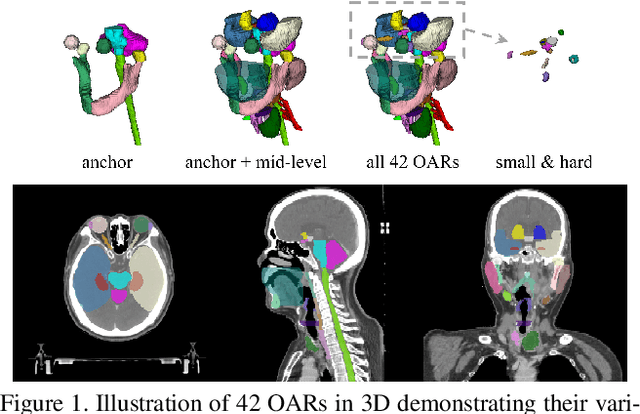
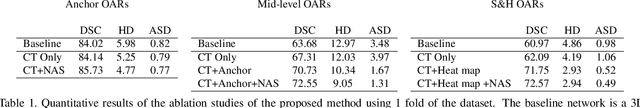
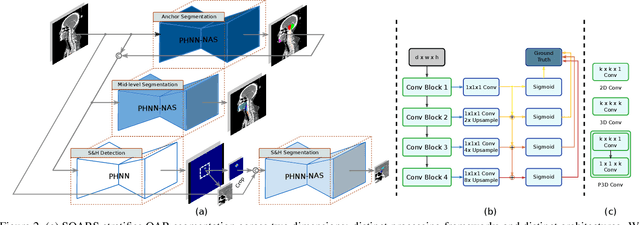
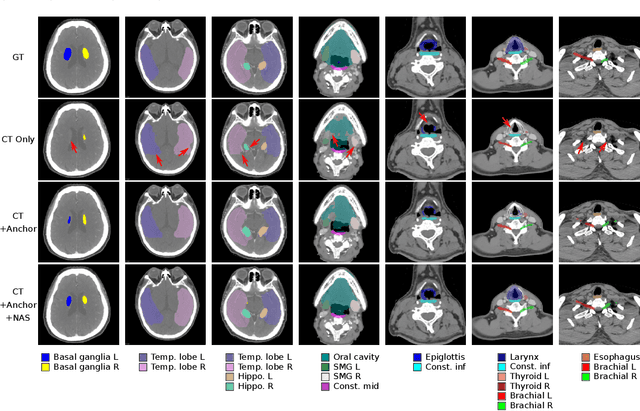
Abstract:OAR segmentation is a critical step in radiotherapy of head and neck (H&N) cancer, where inconsistencies across radiation oncologists and prohibitive labor costs motivate automated approaches. However, leading methods using standard fully convolutional network workflows that are challenged when the number of OARs becomes large, e.g. > 40. For such scenarios, insights can be gained from the stratification approaches seen in manual clinical OAR delineation. This is the goal of our work, where we introduce stratified organ at risk segmentation (SOARS), an approach that stratifies OARs into anchor, mid-level, and small & hard (S&H) categories. SOARS stratifies across two dimensions. The first dimension is that distinct processing pipelines are used for each OAR category. In particular, inspired by clinical practices, anchor OARs are used to guide the mid-level and S&H categories. The second dimension is that distinct network architectures are used to manage the significant contrast, size, and anatomy variations between different OARs. We use differentiable neural architecture search (NAS), allowing the network to choose among 2D, 3D or Pseudo-3D convolutions. Extensive 4-fold cross-validation on 142 H&N cancer patients with 42 manually labeled OARs, the most comprehensive OAR dataset to date, demonstrates that both pipeline- and NAS-stratification significantly improves quantitative performance over the state-of-the-art (from 69.52% to 73.68% in absolute Dice scores). Thus, SOARS provides a powerful and principled means to manage the highly complex segmentation space of OARs.
Radiotherapy Target Contouring with Convolutional Gated Graph Neural Network
Apr 05, 2019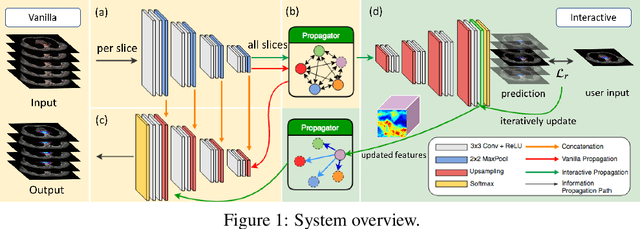
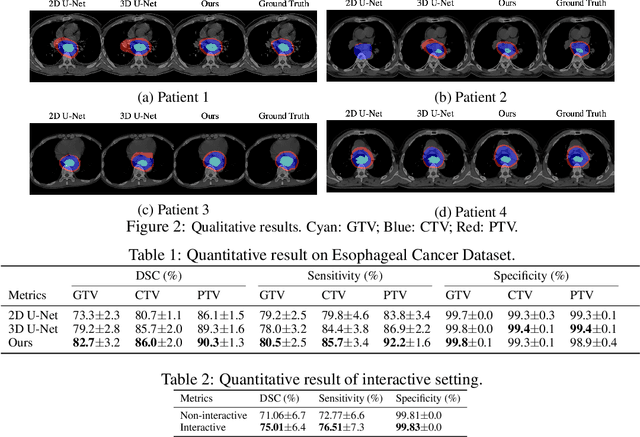
Abstract:Tomography medical imaging is essential in the clinical workflow of modern cancer radiotherapy. Radiation oncologists identify cancerous tissues, applying delineation on treatment regions throughout all image slices. This kind of task is often formulated as a volumetric segmentation task by means of 3D convolutional networks with considerable computational cost. Instead, inspired by the treating methodology of considering meaningful information across slices, we used Gated Graph Neural Network to frame this problem more efficiently. More specifically, we propose convolutional recurrent Gated Graph Propagator (GGP) to propagate high-level information through image slices, with learnable adjacency weighted matrix. Furthermore, as physicians often investigate a few specific slices to refine their decision, we model this slice-wise interaction procedure to further improve our segmentation result. This can be set by editing any slice effortlessly as updating predictions of other slices using GGP. To evaluate our method, we collect an Esophageal Cancer Radiotherapy Target Treatment Contouring dataset of 81 patients which includes tomography images with radiotherapy target. On this dataset, our convolutional graph network produces state-of-the-art results and outperforms the baselines. With the addition of interactive setting, performance is improved even further. Our method has the potential to be easily applied to diverse kinds of medical tasks with volumetric images. Incorporating both the ability to make a feasible prediction and to consider the human interactive input, the proposed method is suitable for clinical scenarios.
Cube Padding for Weakly-Supervised Saliency Prediction in 360° Videos
Jun 04, 2018
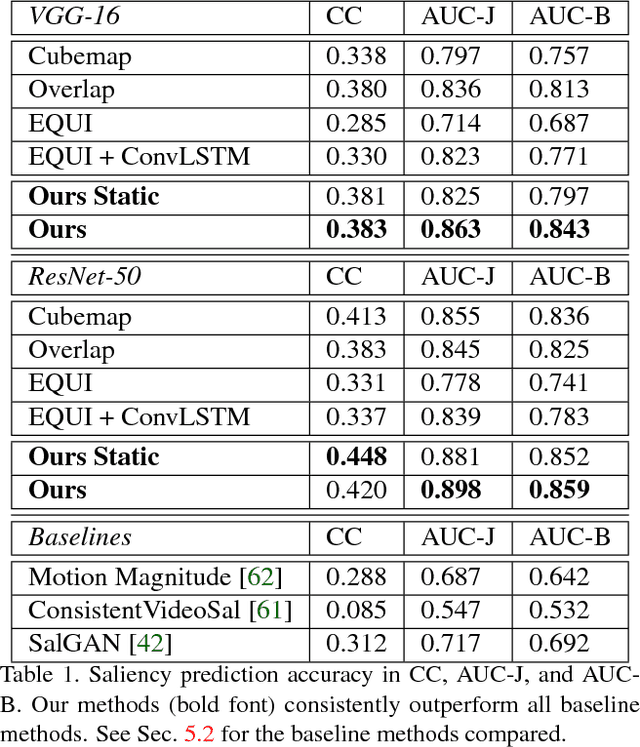

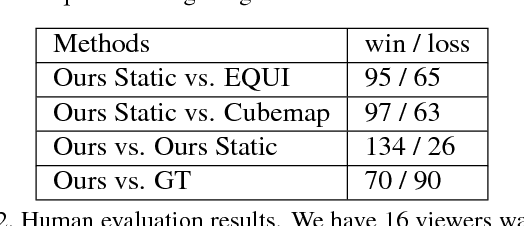
Abstract:Automatic saliency prediction in 360{\deg} videos is critical for viewpoint guidance applications (e.g., Facebook 360 Guide). We propose a spatial-temporal network which is (1) weakly-supervised trained and (2) tailor-made for 360{\deg} viewing sphere. Note that most existing methods are less scalable since they rely on annotated saliency map for training. Most importantly, they convert 360{\deg} sphere to 2D images (e.g., a single equirectangular image or multiple separate Normal Field-of-View (NFoV) images) which introduces distortion and image boundaries. In contrast, we propose a simple and effective Cube Padding (CP) technique as follows. Firstly, we render the 360{\deg} view on six faces of a cube using perspective projection. Thus, it introduces very little distortion. Then, we concatenate all six faces while utilizing the connectivity between faces on the cube for image padding (i.e., Cube Padding) in convolution, pooling, convolutional LSTM layers. In this way, CP introduces no image boundary while being applicable to almost all Convolutional Neural Network (CNN) structures. To evaluate our method, we propose Wild-360, a new 360{\deg} video saliency dataset, containing challenging videos with saliency heatmap annotations. In experiments, our method outperforms baseline methods in both speed and quality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge