Botong Wu
Causal Hidden Markov Model for Time Series Disease Forecasting
Mar 30, 2021



Abstract:We propose a causal hidden Markov model to achieve robust prediction of irreversible disease at an early stage, which is safety-critical and vital for medical treatment in early stages. Specifically, we introduce the hidden variables which propagate to generate medical data at each time step. To avoid learning spurious correlation (e.g., confounding bias), we explicitly separate these hidden variables into three parts: a) the disease (clinical)-related part; b) the disease (non-clinical)-related part; c) others, with only a),b) causally related to the disease however c) may contain spurious correlations (with the disease) inherited from the data provided. With personal attributes and the disease label respectively provided as side information and supervision, we prove that these disease-related hidden variables can be disentangled from others, implying the avoidance of spurious correlation for generalization to medical data from other (out-of-) distributions. Guaranteed by this result, we propose a sequential variational auto-encoder with a reformulated objective function. We apply our model to the early prediction of peripapillary atrophy and achieve promising results on out-of-distribution test data. Further, the ablation study empirically shows the effectiveness of each component in our method. And the visualization shows the accurate identification of lesion regions from others.
Disease Forecast via Progression Learning
Dec 21, 2020



Abstract:Forecasting Parapapillary atrophy (PPA), i.e., a symptom related to most irreversible eye diseases, provides an alarm for implementing an intervention to slow down the disease progression at early stage. A key question for this forecast is: how to fully utilize the historical data (e.g., retinal image) up to the current stage for future disease prediction? In this paper, we provide an answer with a novel framework, namely \textbf{D}isease \textbf{F}orecast via \textbf{P}rogression \textbf{L}earning (\textbf{DFPL}), which exploits the irreversibility prior (i.e., cannot be reversed once diagnosed). Specifically, based on this prior, we decompose two factors that contribute to the prediction of the future disease: i) the current disease label given the data (retinal image, clinical attributes) at present and ii) the future disease label given the progression of the retinal images that from the current to the future. To model these two factors, we introduce the current and progression predictors in DFPL, respectively. In order to account for the degree of progression of the disease, we propose a temporal generative model to accurately generate the future image and compare it with the current one to get a residual image. The generative model is implemented by a recurrent neural network, in order to exploit the dependency of the historical data. To verify our approach, we apply it to a PPA in-house dataset and it yields a significant improvement (\textit{e.g.}, \textbf{4.48\%} of accuracy; \textbf{3.45\%} of AUC) over others. Besides, our generative model can accurately localize the disease-related regions.
Identifying Invariant Texture Violation for Robust Deepfake Detection
Dec 19, 2020



Abstract:Existing deepfake detection methods have reported promising in-distribution results, by accessing published large-scale dataset. However, due to the non-smooth synthesis method, the fake samples in this dataset may expose obvious artifacts (e.g., stark visual contrast, non-smooth boundary), which were heavily relied on by most of the frame-level detection methods above. As these artifacts do not come up in real media forgeries, the above methods can suffer from a large degradation when applied to fake images that close to reality. To improve the robustness for high-realism fake data, we propose the Invariant Texture Learning (InTeLe) framework, which only accesses the published dataset with low visual quality. Our method is based on the prior that the microscopic facial texture of the source face is inevitably violated by the texture transferred from the target person, which can hence be regarded as the invariant characterization shared among all fake images. To learn such an invariance for deepfake detection, our InTeLe introduces an auto-encoder framework with different decoders for pristine and fake images, which are further appended with a shallow classifier in order to separate out the obvious artifact-effect. Equipped with such a separation, the extracted embedding by encoder can capture the texture violation in fake images, followed by the classifier for the final pristine/fake prediction. As a theoretical guarantee, we prove the identifiability of such an invariance texture violation, i.e., to be precisely inferred from observational data. The effectiveness and utility of our method are demonstrated by promising generalization ability from low-quality images with obvious artifacts to fake images with high realism.
Latent Causal Invariant Model
Nov 04, 2020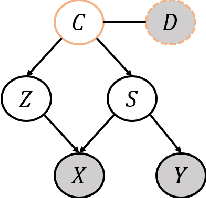
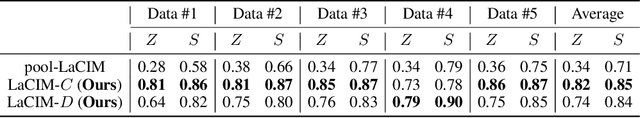
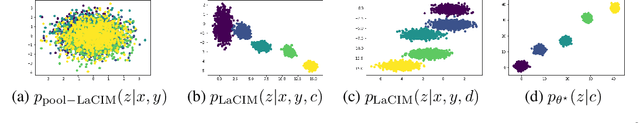
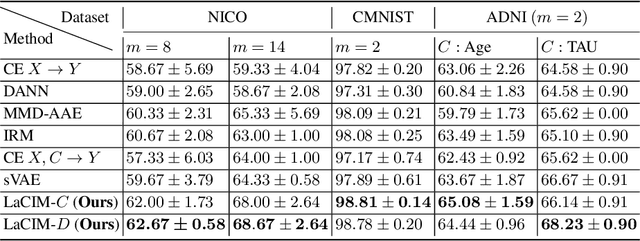
Abstract:Current supervised learning can learn spurious correlation during the data-fitting process, imposing issues regarding interpretability, out-of-distribution (OOD) generalization, and robustness. To avoid spurious correlation, we propose a Latent Causal Invariance Model (LaCIM) which pursues causal prediction. Specifically, we introduce latent variables that are separated into (a) output-causative factors and (b) others that are spuriously correlated to the output via confounders, to model the underlying causal factors. We further assume the generating mechanisms from latent space to observed data to be causally invariant. We give the identifiable claim of such invariance, particularly the disentanglement of output-causative factors from others, as a theoretical guarantee for precise inference and avoiding spurious correlation. We propose a Variational-Bayesian-based method for estimation and to optimize over the latent space for prediction. The utility of our approach is verified by improved interpretability, prediction power on various OOD scenarios (including healthcare) and robustness on security.
Joint Learning for Pulmonary Nodule Segmentation, Attributes and Malignancy Prediction
Feb 10, 2018



Abstract:Refer to the literature of lung nodule classification, many studies adopt Convolutional Neural Networks (CNN) to directly predict the malignancy of lung nodules with original thoracic Computed Tomography (CT) and nodule location. However, these studies cannot tell how the CNN works in terms of predicting the malignancy of the given nodule, e.g., it's hard to conclude that whether the region within the nodule or the contextual information matters according to the output of the CNN. In this paper, we propose an interpretable and multi-task learning CNN -- Joint learning for \textbf{P}ulmonary \textbf{N}odule \textbf{S}egmentation \textbf{A}ttributes and \textbf{M}alignancy \textbf{P}rediction (PN-SAMP). It is able to not only accurately predict the malignancy of lung nodules, but also provide semantic high-level attributes as well as the areas of detected nodules. Moreover, the combination of nodule segmentation, attributes and malignancy prediction is helpful to improve the performance of each single task. In addition, inspired by the fact that radiologists often change window widths and window centers to help to make decision on uncertain nodules, PN-SAMP mixes multiple WW/WC together to gain information for the raw CT input images. To verify the effectiveness of the proposed method, the evaluation is implemented on the public LIDC-IDRI dataset, which is one of the largest dataset for lung nodule malignancy prediction. Experiments indicate that the proposed PN-SAMP achieves significant improvement with respect to lung nodule classification, and promising performance on lung nodule segmentation and attribute learning, compared with the-state-of-the-art methods.
Zero-Shot Learning posed as a Missing Data Problem
Feb 21, 2017



Abstract:This paper presents a method of zero-shot learning (ZSL) which poses ZSL as the missing data problem, rather than the missing label problem. Specifically, most existing ZSL methods focus on learning mapping functions from the image feature space to the label embedding space. Whereas, the proposed method explores a simple yet effective transductive framework in the reverse way \--- our method estimates data distribution of unseen classes in the image feature space by transferring knowledge from the label embedding space. In experiments, our method outperforms the state-of-the-art on two popular datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge