Anastasia Oikonomou
AutoRad-Lung: A Radiomic-Guided Prompting Autoregressive Vision-Language Model for Lung Nodule Malignancy Prediction
Mar 26, 2025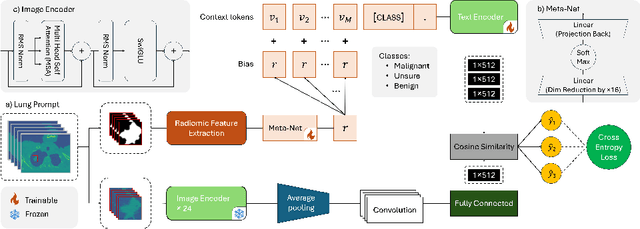
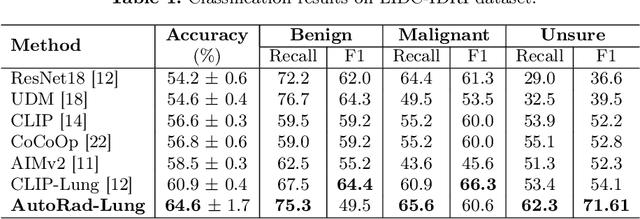
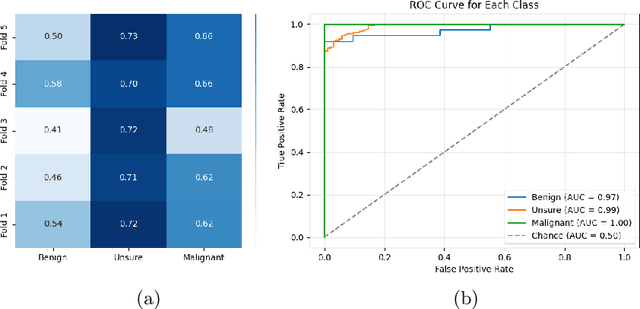
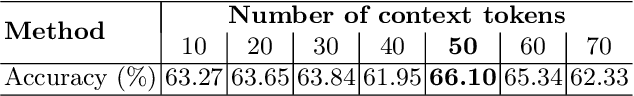
Abstract:Lung cancer remains one of the leading causes of cancer-related mortality worldwide. A crucial challenge for early diagnosis is differentiating uncertain cases with similar visual characteristics and closely annotation scores. In clinical practice, radiologists rely on quantitative, hand-crafted Radiomic features extracted from Computed Tomography (CT) images, while recent research has primarily focused on deep learning solutions. More recently, Vision-Language Models (VLMs), particularly Contrastive Language-Image Pre-Training (CLIP)-based models, have gained attention for their ability to integrate textual knowledge into lung cancer diagnosis. While CLIP-Lung models have shown promising results, we identified the following potential limitations: (a) dependence on radiologists' annotated attributes, which are inherently subjective and error-prone, (b) use of textual information only during training, limiting direct applicability at inference, and (c) Convolutional-based vision encoder with randomly initialized weights, which disregards prior knowledge. To address these limitations, we introduce AutoRad-Lung, which couples an autoregressively pre-trained VLM, with prompts generated from hand-crafted Radiomics. AutoRad-Lung uses the vision encoder of the Large-Scale Autoregressive Image Model (AIMv2), pre-trained using a multi-modal autoregressive objective. Given that lung tumors are typically small, irregularly shaped, and visually similar to healthy tissue, AutoRad-Lung offers significant advantages over its CLIP-based counterparts by capturing pixel-level differences. Additionally, we introduce conditional context optimization, which dynamically generates context-specific prompts based on input Radiomics, improving cross-modal alignment.
NYCTALE: Neuro-Evidence Transformer for Adaptive and Personalized Lung Nodule Invasiveness Prediction
Feb 15, 2024Abstract:Drawing inspiration from the primate brain's intriguing evidence accumulation process, and guided by models from cognitive psychology and neuroscience, the paper introduces the NYCTALE framework, a neuro-inspired and evidence accumulation-based Transformer architecture. The proposed neuro-inspired NYCTALE offers a novel pathway in the domain of Personalized Medicine (PM) for lung cancer diagnosis. In nature, Nyctales are small owls known for their nocturnal behavior, hunting primarily during the darkness of night. The NYCTALE operates in a similarly vigilant manner, i.e., processing data in an evidence-based fashion and making predictions dynamically/adaptively. Distinct from conventional Computed Tomography (CT)-based Deep Learning (DL) models, the NYCTALE performs predictions only when sufficient amount of evidence is accumulated. In other words, instead of processing all or a pre-defined subset of CT slices, for each person, slices are provided one at a time. The NYCTALE framework then computes an evidence vector associated with contribution of each new CT image. A decision is made once the total accumulated evidence surpasses a specific threshold. Preliminary experimental analyses conducted using a challenging in-house dataset comprising 114 subjects. The results are noteworthy, suggesting that NYCTALE outperforms the benchmark accuracy even with approximately 60% less training data on this demanding and small dataset.
Spatio-Temporal Hybrid Fusion of CAE and SWIn Transformers for Lung Cancer Malignancy Prediction
Oct 27, 2022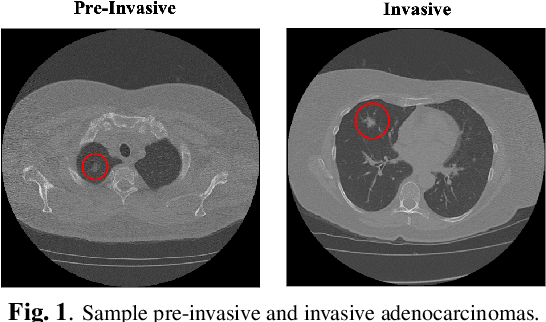
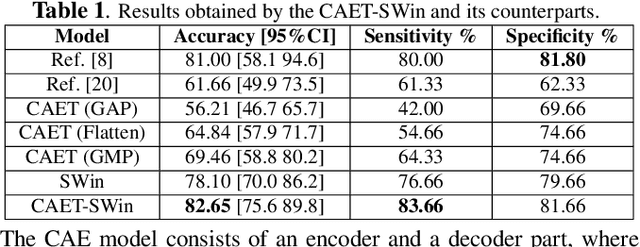
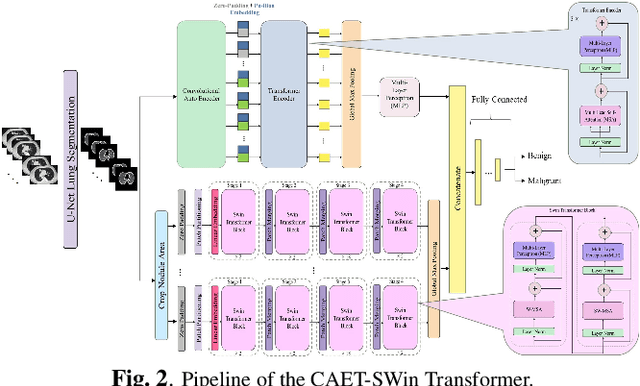
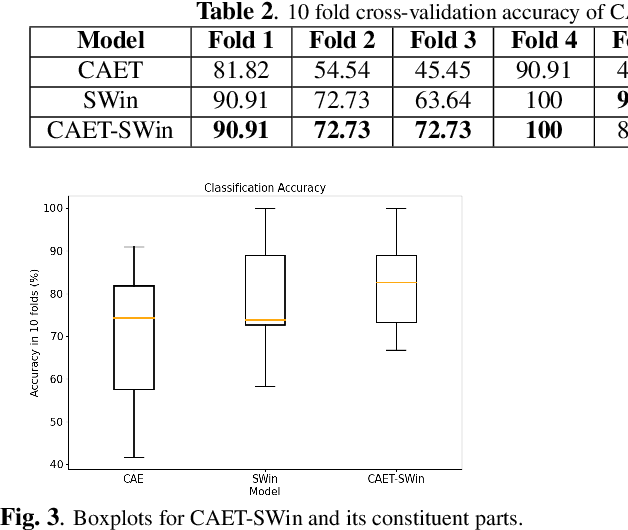
Abstract:The paper proposes a novel hybrid discovery Radiomics framework that simultaneously integrates temporal and spatial features extracted from non-thin chest Computed Tomography (CT) slices to predict Lung Adenocarcinoma (LUAC) malignancy with minimum expert involvement. Lung cancer is the leading cause of mortality from cancer worldwide and has various histologic types, among which LUAC has recently been the most prevalent. LUACs are classified as pre-invasive, minimally invasive, and invasive adenocarcinomas. Timely and accurate knowledge of the lung nodules malignancy leads to a proper treatment plan and reduces the risk of unnecessary or late surgeries. Currently, chest CT scan is the primary imaging modality to assess and predict the invasiveness of LUACs. However, the radiologists' analysis based on CT images is subjective and suffers from a low accuracy compared to the ground truth pathological reviews provided after surgical resections. The proposed hybrid framework, referred to as the CAET-SWin, consists of two parallel paths: (i) The Convolutional Auto-Encoder (CAE) Transformer path that extracts and captures informative features related to inter-slice relations via a modified Transformer architecture, and; (ii) The Shifted Window (SWin) Transformer path, which is a hierarchical vision transformer that extracts nodules' related spatial features from a volumetric CT scan. Extracted temporal (from the CAET-path) and spatial (from the Swin path) are then fused through a fusion path to classify LUACs. Experimental results on our in-house dataset of 114 pathologically proven Sub-Solid Nodules (SSNs) demonstrate that the CAET-SWin significantly improves reliability of the invasiveness prediction task while achieving an accuracy of 82.65%, sensitivity of 83.66%, and specificity of 81.66% using 10-fold cross-validation.
Lung-Originated Tumor Segmentation from Computed Tomography Scan (LOTUS) Benchmark
Jan 03, 2022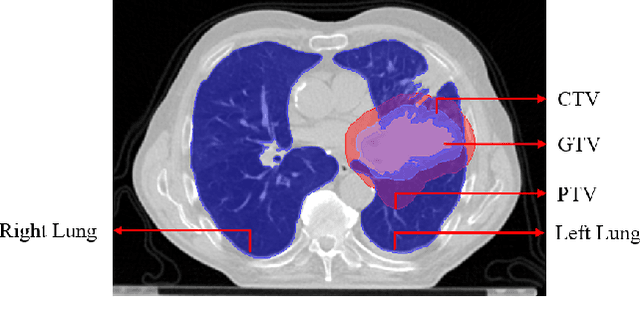
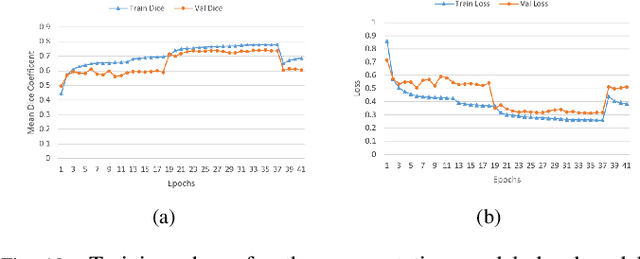
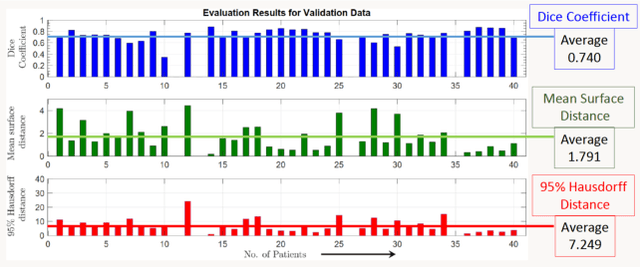
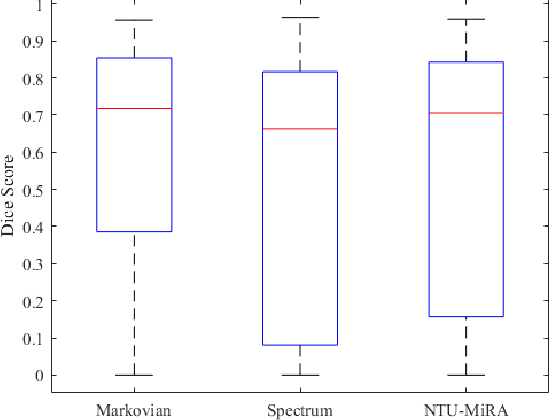
Abstract:Lung cancer is one of the deadliest cancers, and in part its effective diagnosis and treatment depend on the accurate delineation of the tumor. Human-centered segmentation, which is currently the most common approach, is subject to inter-observer variability, and is also time-consuming, considering the fact that only experts are capable of providing annotations. Automatic and semi-automatic tumor segmentation methods have recently shown promising results. However, as different researchers have validated their algorithms using various datasets and performance metrics, reliably evaluating these methods is still an open challenge. The goal of the Lung-Originated Tumor Segmentation from Computed Tomography Scan (LOTUS) Benchmark created through 2018 IEEE Video and Image Processing (VIP) Cup competition, is to provide a unique dataset and pre-defined metrics, so that different researchers can develop and evaluate their methods in a unified fashion. The 2018 VIP Cup started with a global engagement from 42 countries to access the competition data. At the registration stage, there were 129 members clustered into 28 teams from 10 countries, out of which 9 teams made it to the final stage and 6 teams successfully completed all the required tasks. In a nutshell, all the algorithms proposed during the competition, are based on deep learning models combined with a false positive reduction technique. Methods developed by the three finalists show promising results in tumor segmentation, however, more effort should be put into reducing the false positive rate. This competition manuscript presents an overview of the VIP-Cup challenge, along with the proposed algorithms and results.
CAE-Transformer: Transformer-based Model to Predict Invasiveness of Lung Adenocarcinoma Subsolid Nodules from Non-thin Section 3D CT Scans
Oct 17, 2021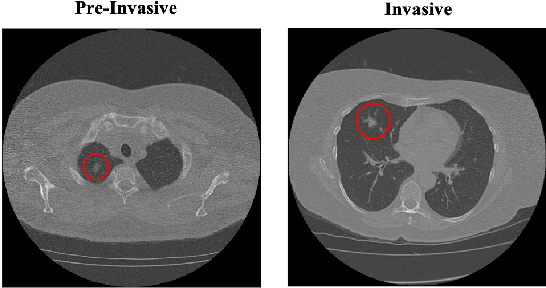
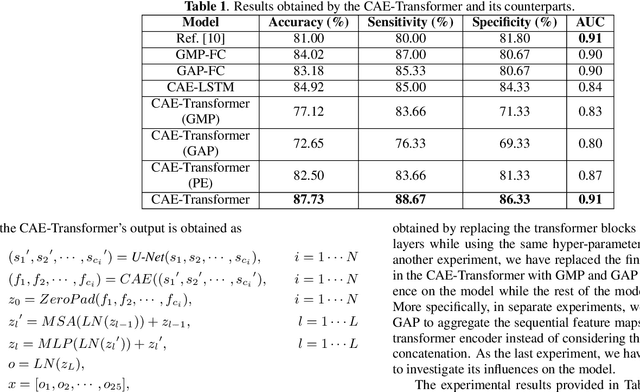
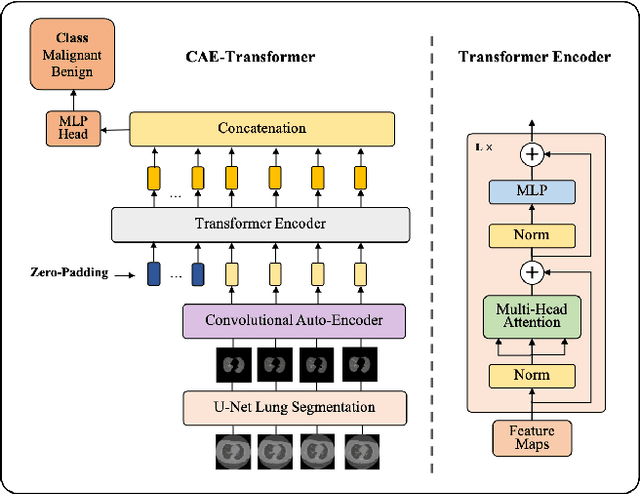
Abstract:Lung cancer is the leading cause of mortality from cancer worldwide and has various histologic types, among which Lung Adenocarcinoma (LAUC) has recently been the most prevalent. Lung adenocarcinomas are classified as pre-invasive, minimally invasive, and invasive adenocarcinomas. Timely and accurate knowledge of the invasiveness of lung nodules leads to a proper treatment plan and reduces the risk of unnecessary or late surgeries. Currently, the primary imaging modality to assess and predict the invasiveness of LAUCs is the chest CT. The results based on CT images, however, are subjective and suffer from a low accuracy compared to the ground truth pathological reviews provided after surgical resections. In this paper, a predictive transformer-based framework, referred to as the "CAE-Transformer", is developed to classify LAUCs. The CAE-Transformer utilizes a Convolutional Auto-Encoder (CAE) to automatically extract informative features from CT slices, which are then fed to a modified transformer model to capture global inter-slice relations. Experimental results on our in-house dataset of 114 pathologically proven Sub-Solid Nodules (SSNs) demonstrate the superiority of the CAE-Transformer over the histogram/radiomics-based models and its deep learning-based counterparts, achieving an accuracy of 87.73%, sensitivity of 88.67%, specificity of 86.33%, and AUC of 0.913, using a 10-fold cross-validation.
Robust Automated Framework for COVID-19 Disease Identification from a Multicenter Dataset of Chest CT Scans
Sep 26, 2021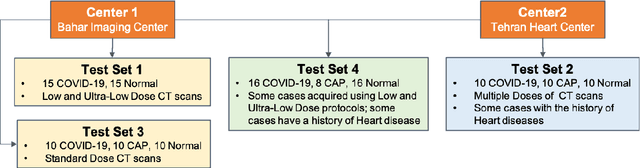
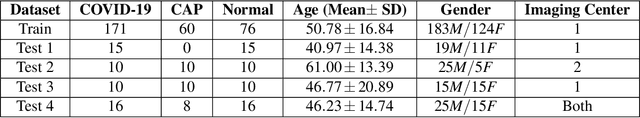
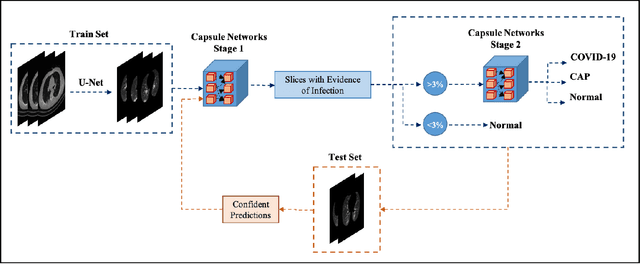
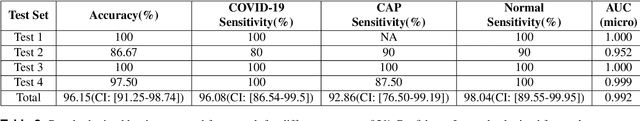
Abstract:The objective of this study is to develop a robust deep learning-based framework to distinguish COVID-19, Community-Acquired Pneumonia (CAP), and Normal cases based on chest CT scans acquired in different imaging centers using various protocols, and radiation doses. We showed that while our proposed model is trained on a relatively small dataset acquired from only one imaging center using a specific scanning protocol, the model performs well on heterogeneous test sets obtained by multiple scanners using different technical parameters. We also showed that the model can be updated via an unsupervised approach to cope with the data shift between the train and test sets and enhance the robustness of the model upon receiving a new external dataset from a different center. We adopted an ensemble architecture to aggregate the predictions from multiple versions of the model. For initial training and development purposes, an in-house dataset of 171 COVID-19, 60 CAP, and 76 Normal cases was used, which contained volumetric CT scans acquired from one imaging center using a constant standard radiation dose scanning protocol. To evaluate the model, we collected four different test sets retrospectively to investigate the effects of the shifts in the data characteristics on the model's performance. Among the test cases, there were CT scans with similar characteristics as the train set as well as noisy low-dose and ultra-low dose CT scans. In addition, some test CT scans were obtained from patients with a history of cardiovascular diseases or surgeries. The entire test dataset used in this study contained 51 COVID-19, 28 CAP, and 51 Normal cases. Experimental results indicate that our proposed framework performs well on all test sets achieving total accuracy of 96.15% (95%CI: [91.25-98.74]), COVID-19 sensitivity of 96.08% (95%CI: [86.54-99.5]), CAP sensitivity of 92.86% (95%CI: [76.50-99.19]).
COVID-Rate: An Automated Framework for Segmentation of COVID-19 Lesions from Chest CT Scans
Jul 04, 2021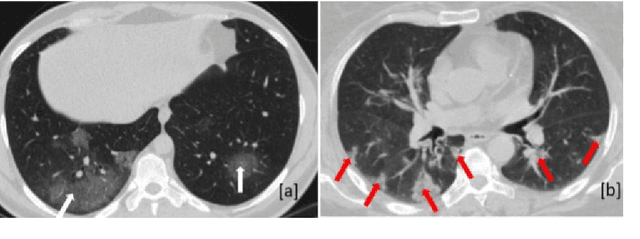

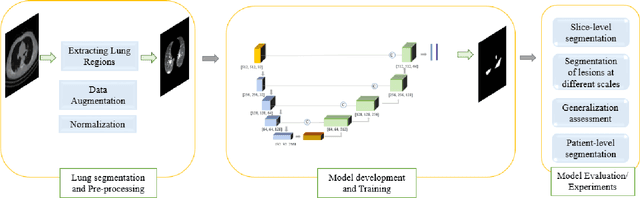
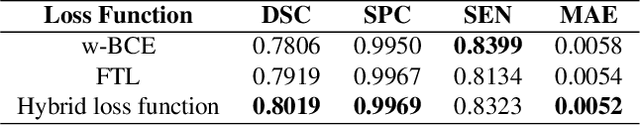
Abstract:Novel Coronavirus disease (COVID-19) is a highly contagious respiratory infection that has had devastating effects on the world. Recently, new COVID-19 variants are emerging making the situation more challenging and threatening. Evaluation and quantification of COVID-19 lung abnormalities based on chest Computed Tomography (CT) scans can help determining the disease stage, efficiently allocating limited healthcare resources, and making informed treatment decisions. During pandemic era, however, visual assessment and quantification of COVID-19 lung lesions by expert radiologists become expensive and prone to error, which raises an urgent quest to develop practical autonomous solutions. In this context, first, the paper introduces an open access COVID-19 CT segmentation dataset containing 433 CT images from 82 patients that have been annotated by an expert radiologist. Second, a Deep Neural Network (DNN)-based framework is proposed, referred to as the COVID-Rate, that autonomously segments lung abnormalities associated with COVID-19 from chest CT scans. Performance of the proposed COVID-Rate framework is evaluated through several experiments based on the introduced and external datasets. The results show a dice score of 0:802 and specificity and sensitivity of 0:997 and 0:832, respectively. Furthermore, the results indicate that the COVID-Rate model can efficiently segment COVID-19 lesions in both 2D CT images and whole lung volumes. Results on the external dataset illustrate generalization capabilities of the COVID-Rate model to CT images obtained from a different scanner.
Human-level COVID-19 Diagnosis from Low-dose CT Scans Using a Two-stage Time-distributed Capsule Network
May 31, 2021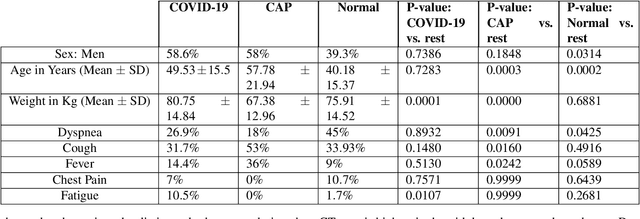
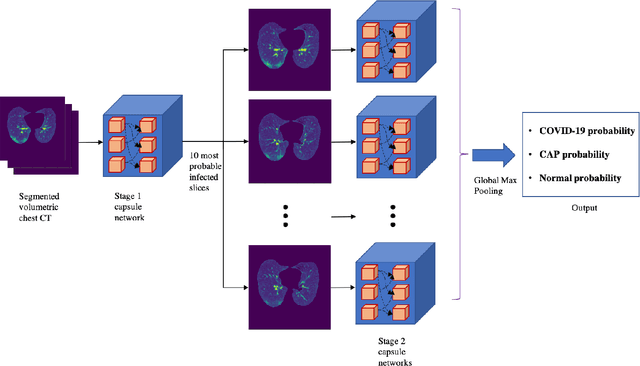
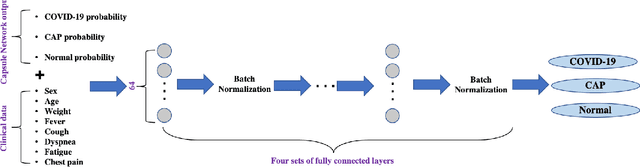
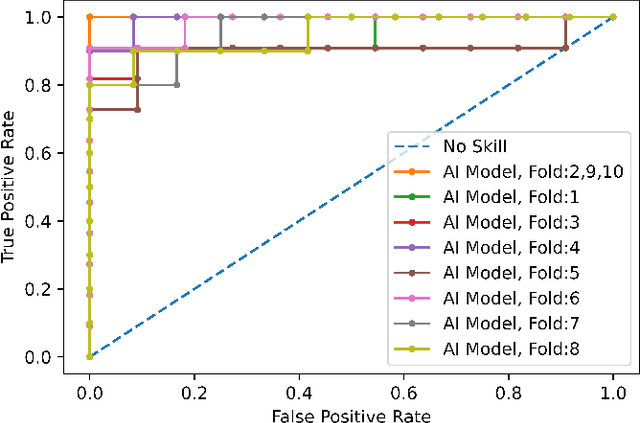
Abstract:Reverse transcription-polymerase chain reaction (RT-PCR) is currently the gold standard in COVID-19 diagnosis. It can, however, take days to provide the diagnosis, and false negative rate is relatively high. Imaging, in particular chest computed tomography (CT), can assist with diagnosis and assessment of this disease. Nevertheless, it is shown that standard dose CT scan gives significant radiation burden to patients, especially those in need of multiple scans. In this study, we consider low-dose and ultra-low-dose (LDCT and ULDCT) scan protocols that reduce the radiation exposure close to that of a single X-Ray, while maintaining an acceptable resolution for diagnosis purposes. Since thoracic radiology expertise may not be widely available during the pandemic, we develop an Artificial Intelligence (AI)-based framework using a collected dataset of LDCT/ULDCT scans, to study the hypothesis that the AI model can provide human-level performance. The AI model uses a two stage capsule network architecture and can rapidly classify COVID-19, community acquired pneumonia (CAP), and normal cases, using LDCT/ULDCT scans. The AI model achieves COVID-19 sensitivity of 89.5% +\- 0.11, CAP sensitivity of 95% +\- 0.11, normal cases sensitivity (specificity) of 85.7% +\- 0.16, and accuracy of 90% +\- 0.06. By incorporating clinical data (demographic and symptoms), the performance further improves to COVID-19 sensitivity of 94.3% +\- pm 0.05, CAP sensitivity of 96.7% +\- 0.07, normal cases sensitivity (specificity) of 91% +\- 0.09 , and accuracy of 94.1% +\- 0.03. The proposed AI model achieves human-level diagnosis based on the LDCT/ULDCT scans with reduced radiation exposure. We believe that the proposed AI model has the potential to assist the radiologists to accurately and promptly diagnose COVID-19 infection and help control the transmission chain during the pandemic.
Diagnosis/Prognosis of COVID-19 Images: Challenges, Opportunities, and Applications
Dec 28, 2020
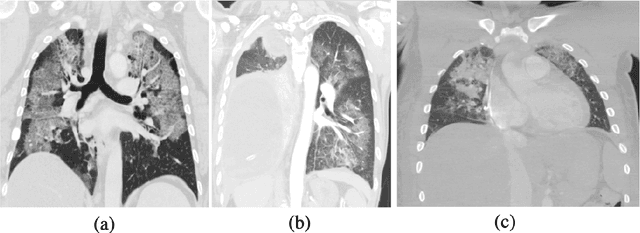
Abstract:The novel Coronavirus disease, COVID-19, has rapidly and abruptly changed the world as we knew in 2020. It becomes the most unprecedent challenge to analytic epidemiology in general and signal processing theories in specific. Given its high contingency nature and adverse effects across the world, it is important to develop efficient processing/learning models to overcome this pandemic and be prepared for potential future ones. In this regard, medical imaging plays an important role for the management of COVID-19. Human-centered interpretation of medical images is, however, tedious and can be subjective. This has resulted in a surge of interest to develop Radiomics models for analysis and interpretation of medical images. Signal Processing (SP) and Deep Learning (DL) models can assist in development of robust Radiomics solutions for diagnosis/prognosis, severity assessment, treatment response, and monitoring of COVID-19 patients. In this article, we aim to present an overview of the current state, challenges, and opportunities of developing SP/DL-empowered models for diagnosis (screening/monitoring) and prognosis (outcome prediction and severity assessment) of COVID-19 infection. More specifically, the article starts by elaborating the latest development on the theoretical framework of analytic epidemiology and hypersignal processing for COVID-19. Afterwards, imaging modalities and Radiological characteristics of COVID-19 are discussed. SL/DL-based Radiomic models specific to the analysis of COVID-19 infection are then described covering the following four domains: Segmentation of COVID-19 lesions; Predictive models for outcome prediction; Severity assessment, and; Diagnosis/classification models. Finally, open problems and opportunities are presented in detail.
CT-CAPS: Feature Extraction-based Automated Framework for COVID-19 Disease Identification from Chest CT Scans using Capsule Networks
Oct 30, 2020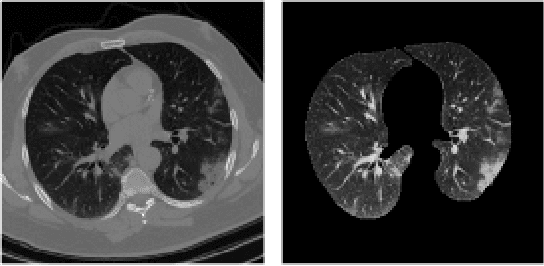
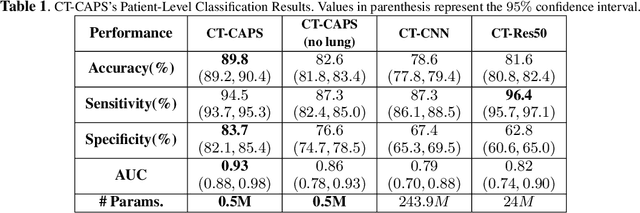
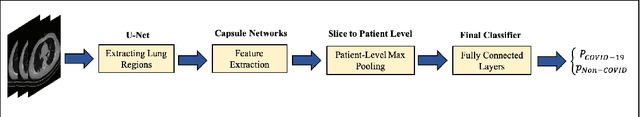
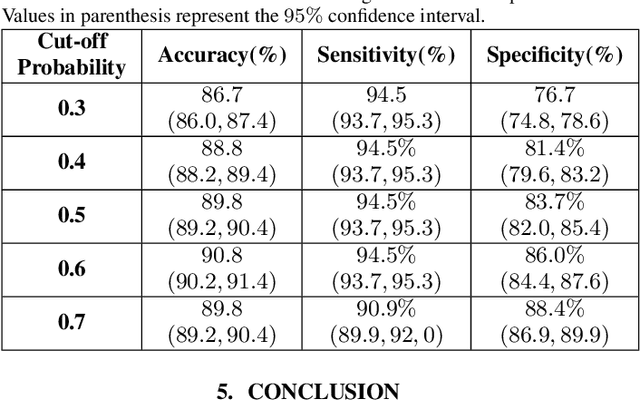
Abstract:The global outbreak of the novel corona virus (COVID-19) disease has drastically impacted the world and led to one of the most challenging crisis across the globe since World War II. The early diagnosis and isolation of COVID-19 positive cases are considered as crucial steps towards preventing the spread of the disease and flattening the epidemic curve. Chest Computed Tomography (CT) scan is a highly sensitive, rapid, and accurate diagnostic technique that can complement Reverse Transcription Polymerase Chain Reaction (RT-PCR) test. Recently, deep learning-based models, mostly based on Convolutional Neural Networks (CNN), have shown promising diagnostic results. CNNs, however, are incapable of capturing spatial relations between image instances and require large datasets. Capsule Networks, on the other hand, can capture spatial relations, require smaller datasets, and have considerably fewer parameters. In this paper, a Capsule network framework, referred to as the "CT-CAPS", is presented to automatically extract distinctive features of chest CT scans. These features, which are extracted from the layer before the final capsule layer, are then leveraged to differentiate COVID-19 from Non-COVID cases. The experiments on our in-house dataset of 307 patients show the state-of-the-art performance with the accuracy of 90.8%, sensitivity of 94.5%, and specificity of 86.0%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge