Andre Dekker
Department of Radiation Oncology
MRI-based and metabolomics-based age scores act synergetically for mortality prediction shown by multi-cohort federated learning
Sep 02, 2024



Abstract:Biological age scores are an emerging tool to characterize aging by estimating chronological age based on physiological biomarkers. Various scores have shown associations with aging-related outcomes. This study assessed the relation between an age score based on brain MRI images (BrainAge) and an age score based on metabolomic biomarkers (MetaboAge). We trained a federated deep learning model to estimate BrainAge in three cohorts. The federated BrainAge model yielded significantly lower error for age prediction across the cohorts than locally trained models. Harmonizing the age interval between cohorts further improved BrainAge accuracy. Subsequently, we compared BrainAge with MetaboAge using federated association and survival analyses. The results showed a small association between BrainAge and MetaboAge as well as a higher predictive value for the time to mortality of both scores combined than for the individual scores. Hence, our study suggests that both aging scores capture different aspects of the aging process.
Federated Bayesian Network Ensembles
Feb 19, 2024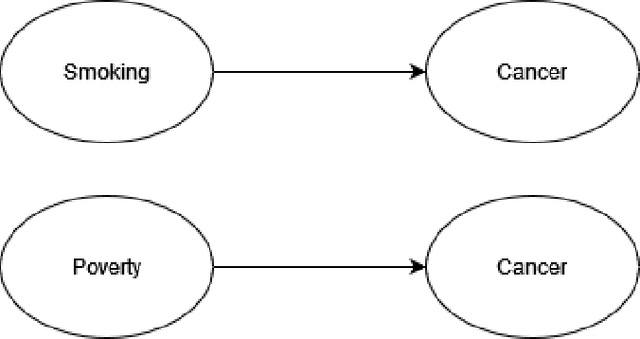
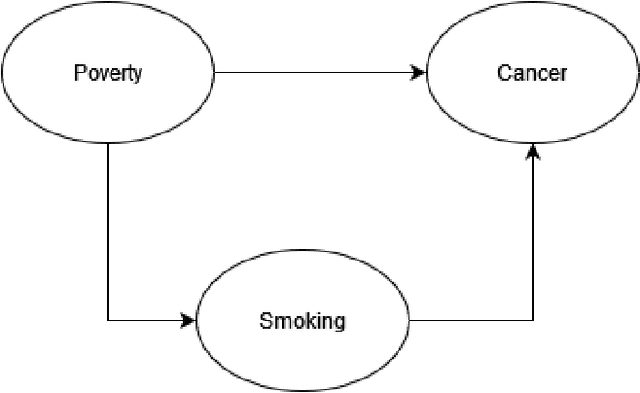
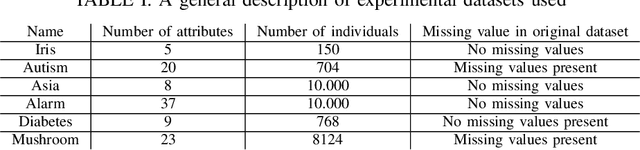
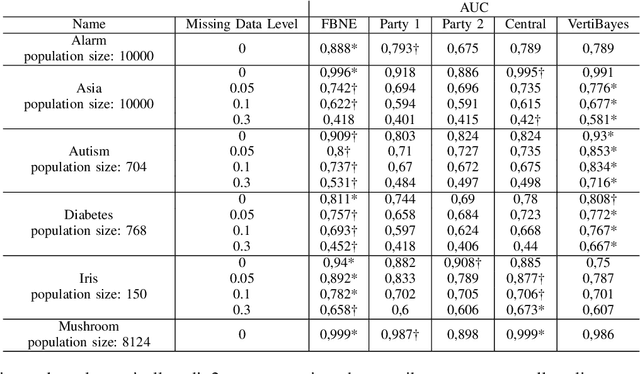
Abstract:Federated learning allows us to run machine learning algorithms on decentralized data when data sharing is not permitted due to privacy concerns. Ensemble-based learning works by training multiple (weak) classifiers whose output is aggregated. Federated ensembles are ensembles applied to a federated setting, where each classifier in the ensemble is trained on one data location. In this article, we explore the use of federated ensembles of Bayesian networks (FBNE) in a range of experiments and compare their performance with locally trained models and models trained with VertiBayes, a federated learning algorithm to train Bayesian networks from decentralized data. Our results show that FBNE outperforms local models and provides a significant increase in training speed compared with VertiBayes while maintaining a similar performance in most settings, among other advantages. We show that FBNE is a potentially useful tool within the federated learning toolbox, especially when local populations are heavily biased, or there is a strong imbalance in population size across parties. We discuss the advantages and disadvantages of this approach in terms of time complexity, model accuracy, privacy protection, and model interpretability.
VertiBayes: Learning Bayesian network parameters from vertically partitioned data with missing values
Oct 31, 2022Abstract:Federated learning makes it possible to train a machine learning model on decentralized data. Bayesian networks are probabilistic graphical models that have been widely used in artificial intelligence applications. Their popularity stems from the fact they can be built by combining existing expert knowledge with data and are highly interpretable, which makes them useful for decision support, e.g. in healthcare. While some research has been published on the federated learning of Bayesian networks, publications on Bayesian networks in a vertically partitioned or heterogeneous data setting (where different variables are located in different datasets) are limited, and suffer from important omissions, such as the handling of missing data. In this article, we propose a novel method called VertiBayes to train Bayesian networks (structure and parameters) on vertically partitioned data, which can handle missing values as well as an arbitrary number of parties. For structure learning we adapted the widely used K2 algorithm with a privacy-preserving scalar product protocol. For parameter learning, we use a two-step approach: first, we learn an intermediate model using maximum likelihood by treating missing values as a special value and then we train a model on synthetic data generated by the intermediate model using the EM algorithm. The privacy guarantees of our approach are equivalent to the ones provided by the privacy preserving scalar product protocol used. We experimentally show our approach produces models comparable to those learnt using traditional algorithms and we estimate the increase in complexity in terms of samples, network size, and complexity. Finally, we propose two alternative approaches to estimate the performance of the model using vertically partitioned data and we show in experiments that they lead to reasonably accurate estimates.
AI Technical Considerations: Data Storage, Cloud usage and AI Pipeline
Jan 20, 2022Abstract:Artificial intelligence (AI), especially deep learning, requires vast amounts of data for training, testing, and validation. Collecting these data and the corresponding annotations requires the implementation of imaging biobanks that provide access to these data in a standardized way. This requires careful design and implementation based on the current standards and guidelines and complying with the current legal restrictions. However, the realization of proper imaging data collections is not sufficient to train, validate and deploy AI as resource demands are high and require a careful hybrid implementation of AI pipelines both on-premise and in the cloud. This chapter aims to help the reader when technical considerations have to be made about the AI environment by providing a technical background of different concepts and implementation aspects involved in data storage, cloud usage, and AI pipelines.
Lung-Originated Tumor Segmentation from Computed Tomography Scan (LOTUS) Benchmark
Jan 03, 2022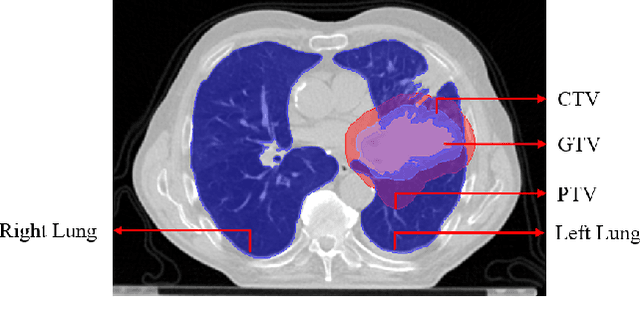
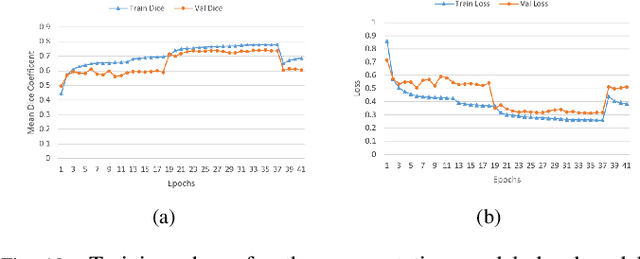
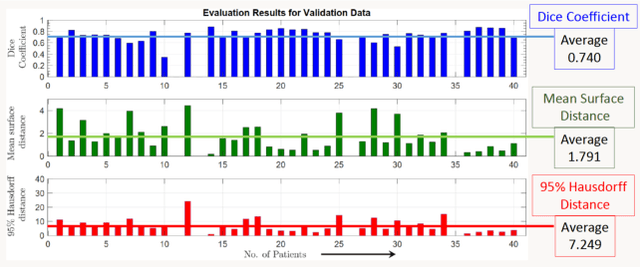
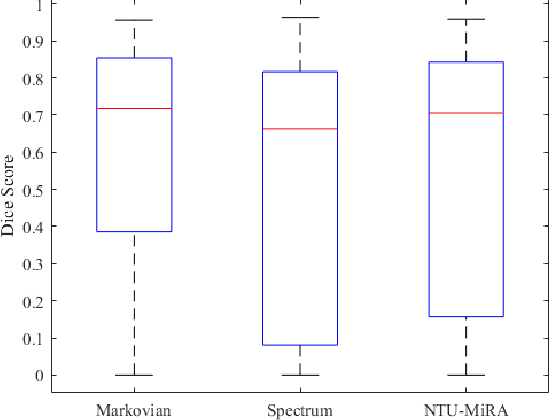
Abstract:Lung cancer is one of the deadliest cancers, and in part its effective diagnosis and treatment depend on the accurate delineation of the tumor. Human-centered segmentation, which is currently the most common approach, is subject to inter-observer variability, and is also time-consuming, considering the fact that only experts are capable of providing annotations. Automatic and semi-automatic tumor segmentation methods have recently shown promising results. However, as different researchers have validated their algorithms using various datasets and performance metrics, reliably evaluating these methods is still an open challenge. The goal of the Lung-Originated Tumor Segmentation from Computed Tomography Scan (LOTUS) Benchmark created through 2018 IEEE Video and Image Processing (VIP) Cup competition, is to provide a unique dataset and pre-defined metrics, so that different researchers can develop and evaluate their methods in a unified fashion. The 2018 VIP Cup started with a global engagement from 42 countries to access the competition data. At the registration stage, there were 129 members clustered into 28 teams from 10 countries, out of which 9 teams made it to the final stage and 6 teams successfully completed all the required tasks. In a nutshell, all the algorithms proposed during the competition, are based on deep learning models combined with a false positive reduction technique. Methods developed by the three finalists show promising results in tumor segmentation, however, more effort should be put into reducing the false positive rate. This competition manuscript presents an overview of the VIP-Cup challenge, along with the proposed algorithms and results.
Predicting erectile dysfunction after treatment for localized prostate cancer
Oct 01, 2021



Abstract:While the 10-year survival rate for localized prostate cancer patients is very good (>98%), side effects of treatment may limit quality of life significantly. Erectile dysfunction (ED) is a common burden associated with increasing age as well as prostate cancer treatment. Although many studies have investigated the factors affecting erectile dysfunction (ED) after prostate cancer treatment, only limited studies have investigated whether ED can be predicted before the start of treatment. The advent of machine learning (ML) based prediction tools in oncology offers a promising approach to improve accuracy of prediction and quality of care. Predicting ED may help aid shared decision making by making the advantages and disadvantages of certain treatments clear, so that a tailored treatment for an individual patient can be chosen. This study aimed to predict ED at 1-year and 2-year post-diagnosis based on patient demographics, clinical data and patient-reported outcomes (PROMs) measured at diagnosis.
Generative Models Improve Radiomics Performance in Different Tasks and Different Datasets: An Experimental Study
Sep 06, 2021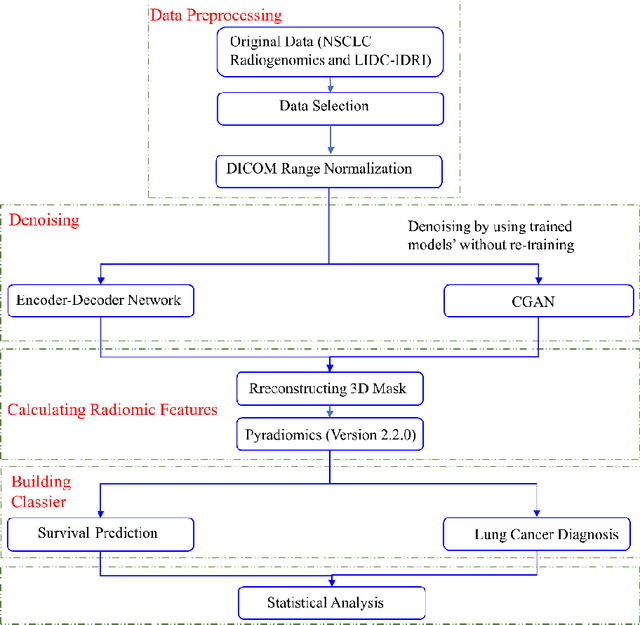
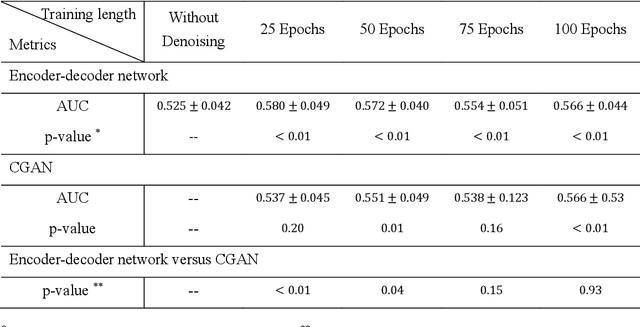
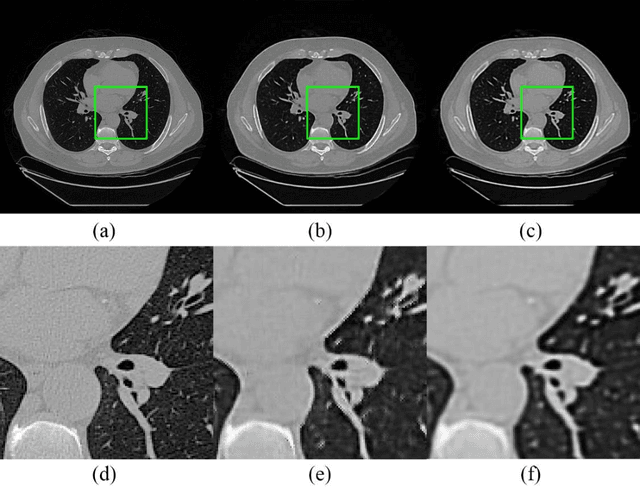
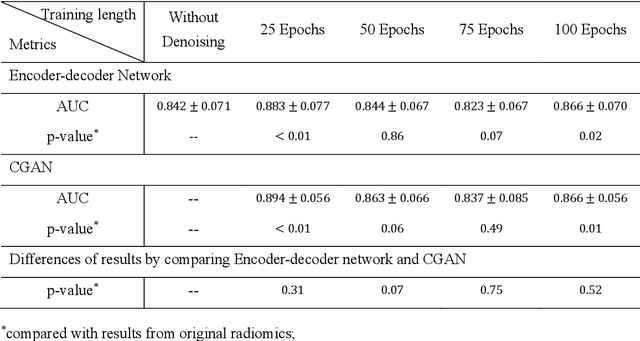
Abstract:Radiomics is an active area of research focusing on high throughput feature extraction from medical images with a wide array of applications in clinical practice, such as clinical decision support in oncology. However, noise in low dose computed tomography (CT) scans can impair the accurate extraction of radiomic features. In this article, we investigate the possibility of using deep learning generative models to improve the performance of radiomics from low dose CTs. We used two datasets of low dose CT scans -NSCLC Radiogenomics and LIDC-IDRI - as test datasets for two tasks - pre-treatment survival prediction and lung cancer diagnosis. We used encoder-decoder networks and conditional generative adversarial networks (CGANs) trained in a previous study as generative models to transform low dose CT images into full dose CT images. Radiomic features extracted from the original and improved CT scans were used to build two classifiers - a support vector machine (SVM) and a deep attention based multiple instance learning model - for survival prediction and lung cancer diagnosis respectively. Finally, we compared the performance of the models derived from the original and improved CT scans. Encoder-decoder networks and CGANs improved the area under the curve (AUC) of survival prediction from 0.52 to 0.57 (p-value<0.01). On the other hand, Encoder-decoder network and CGAN can improve the AUC of lung cancer diagnosis from 0.84 to 0.88 and 0.89 respectively (p-value<0.01). Moreover, there are no statistically significant differences in improving AUC by using encoder-decoder network and CGAN (p-value=0.34) when networks trained at 75 and 100 epochs. Generative models can improve the performance of low dose CT-based radiomics in different tasks. Hence, denoising using generative models seems to be a necessary pre-processing step for calculating radiomic features from low dose CTs.
Generative Models Improve Radiomics Reproducibility in Low Dose CTs: A Simulation Study
Apr 30, 2021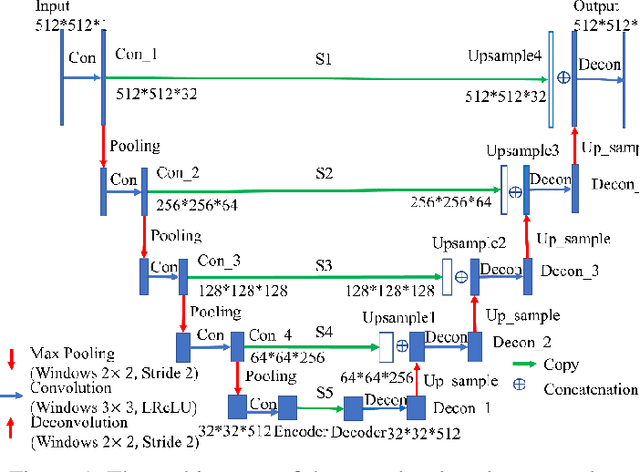
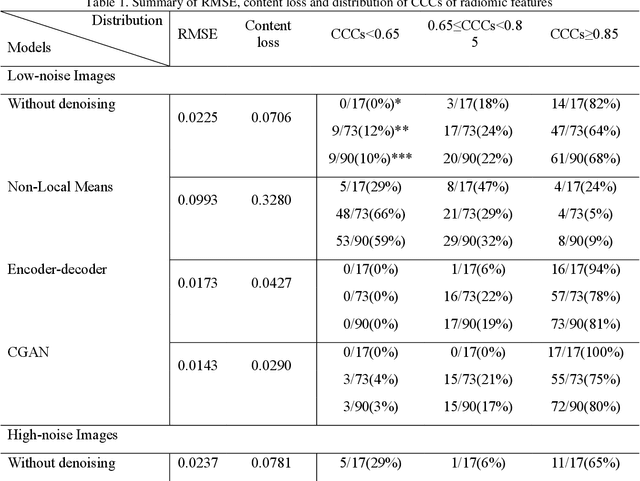
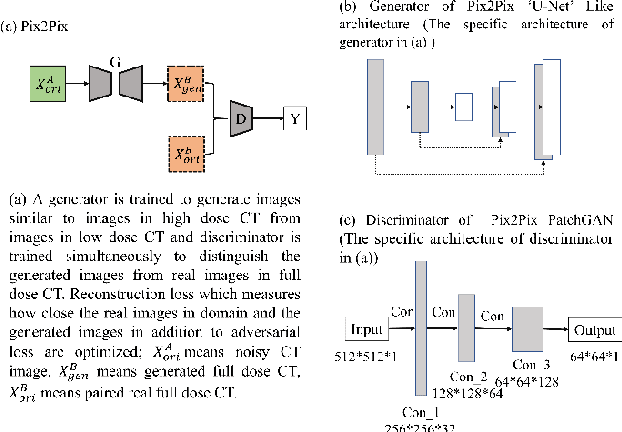
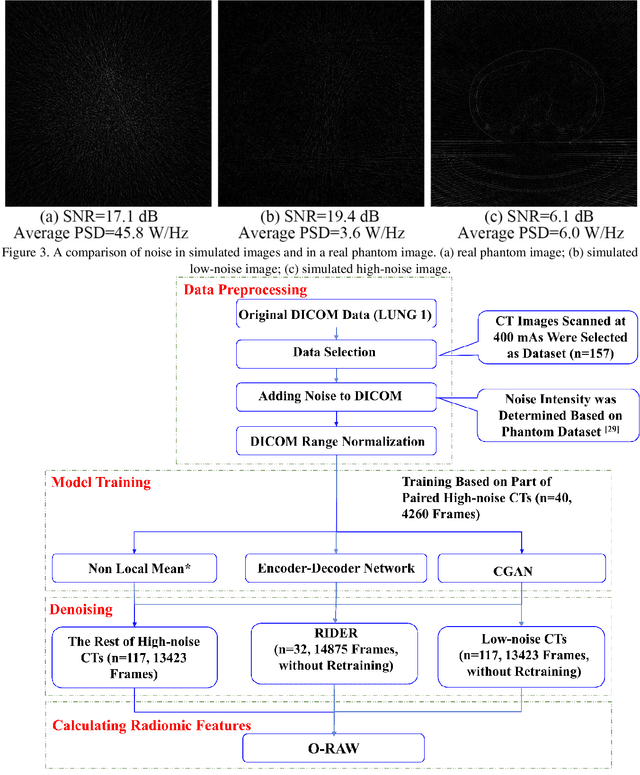
Abstract:Radiomics is an active area of research in medical image analysis, the low reproducibility of radiomics has limited its applicability to clinical practice. This issue is especially prominent when radiomic features are calculated from noisy images, such as low dose computed tomography (CT) scans. In this article, we investigate the possibility of improving the reproducibility of radiomic features calculated on noisy CTs by using generative models for denoising.One traditional denoising method - non-local means - and two generative models - encoder-decoder networks (EDN) and conditional generative adversarial networks (CGANs) - were selected as the test models. We added noise to the sinograms of full dose CTs to mimic low dose CTs with two different levels of noise: low-noise CT and high-noise CT. Models were trained on high-noise CTs and used to denoise low-noise CTs without re-training. We also test the performance of our model in real data, using dataset of same-day repeat low dose CTs to assess the reproducibility of radiomic features in denoised images. The EDN and the CGAN improved the concordance correlation coefficients (CCC) of radiomic features for low-noise images from 0.87 to 0.92 and for high-noise images from 0.68 to 0.92 respectively. Moreover, the EDN and the CGAN improved the test-retest reliability of radiomic features (mean CCC increased from 0.89 to 0.94) based on real low dose CTs. The results show that denoising using EDN and CGANs can improve the reproducibility of radiomic features calculated on noisy CTs. Moreover, images with different noise levels can be denoised to improve the reproducibility using these models without re-training, as long as the noise intensity is equal or lower than that in high-noise CTs. To the authors' knowledge, this is the first effort to improve the reproducibility of radiomic features calculated on low dose CT scans.
Lung Cancer Diagnosis Using Deep Attention Based on Multiple Instance Learning and Radiomics
Apr 29, 2021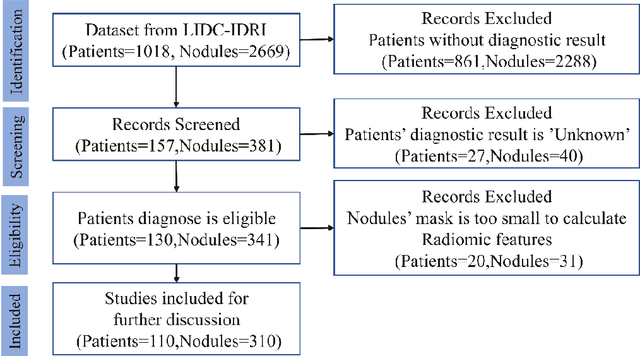
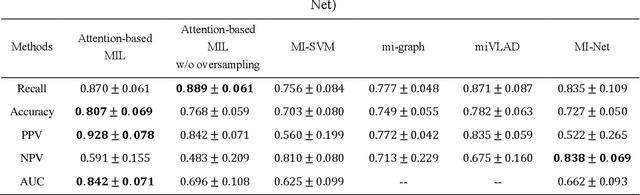
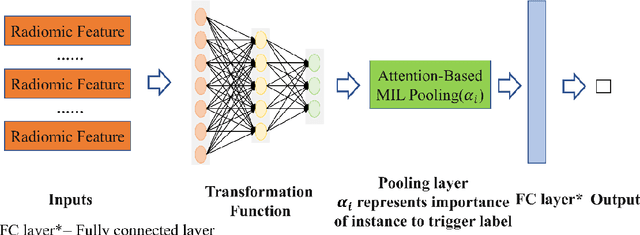
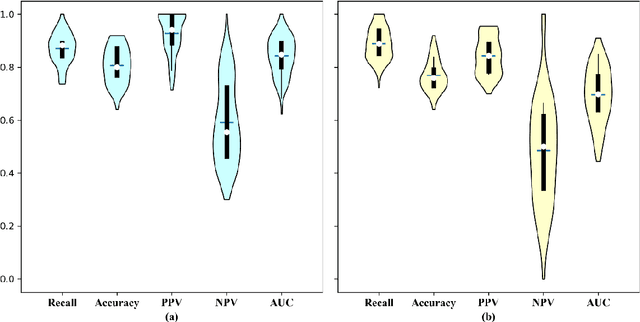
Abstract:Early diagnosis of lung cancer is a key intervention for the treatment of lung cancer computer aided diagnosis (CAD) can play a crucial role. However, most published CAD methods treat lung cancer diagnosis as a lung nodule classification problem, which does not reflect clinical practice, where clinicians diagnose a patient based on a set of images of nodules, instead of one specific nodule. Besides, the low interpretability of the output provided by these methods presents an important barrier for their adoption. In this article, we treat lung cancer diagnosis as a multiple instance learning (MIL) problem in order to better reflect the diagnosis process in the clinical setting and for the higher interpretability of the output. We chose radiomics as the source of input features and deep attention-based MIL as the classification algorithm.The attention mechanism provides higher interpretability by estimating the importance of each instance in the set for the final diagnosis.In order to improve the model's performance in a small imbalanced dataset, we introduce a new bag simulation method for MIL.The results show that our method can achieve a mean accuracy of 0.807 with a standard error of the mean (SEM) of 0.069, a recall of 0.870 (SEM 0.061), a positive predictive value of 0.928 (SEM 0.078), a negative predictive value of 0.591 (SEM 0.155) and an area under the curve (AUC) of 0.842 (SEM 0.074), outperforming other MIL methods.Additional experiments show that the proposed oversampling strategy significantly improves the model's performance. In addition, our experiments show that our method provides an indication of the importance of each nodule in determining the diagnosis, which combined with the well-defined radiomic features, make the results more interpretable and acceptable for doctors and patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge