Alberto Traverso
Generative Models Improve Radiomics Reproducibility in Low Dose CTs: A Simulation Study
Apr 30, 2021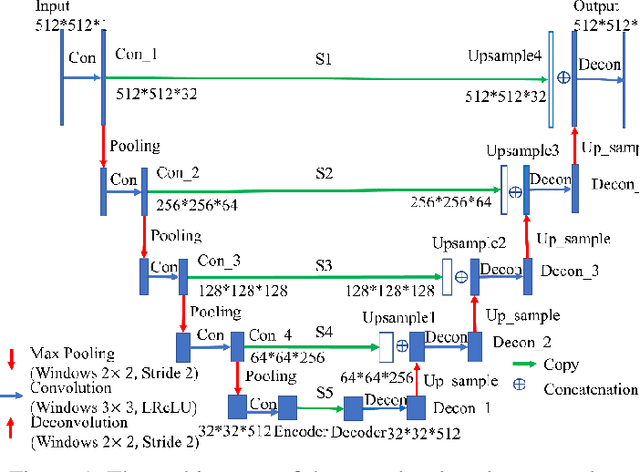
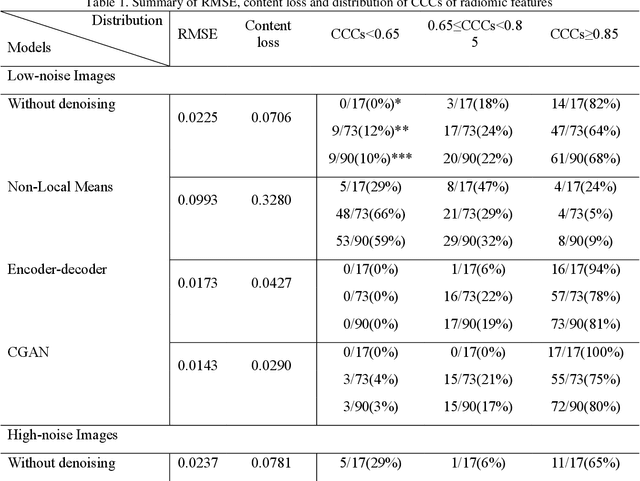
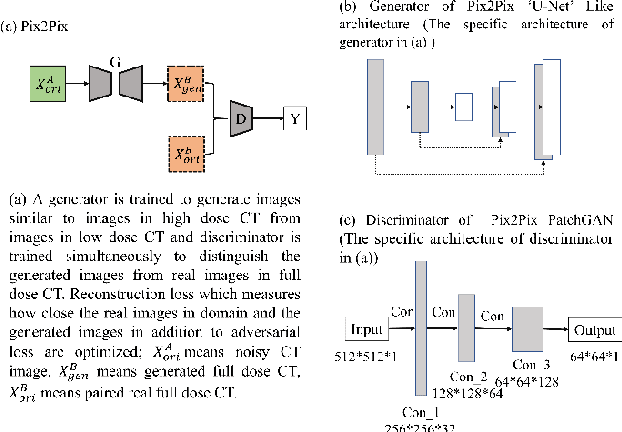
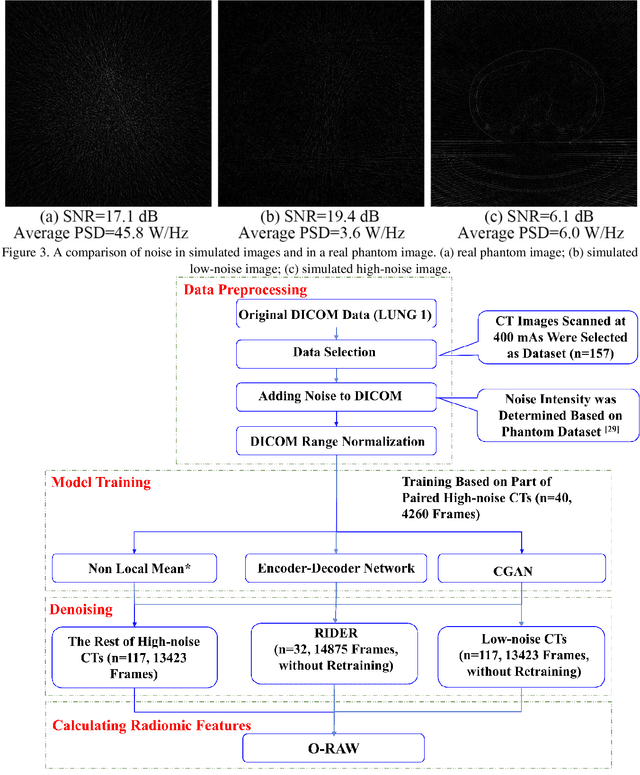
Abstract:Radiomics is an active area of research in medical image analysis, the low reproducibility of radiomics has limited its applicability to clinical practice. This issue is especially prominent when radiomic features are calculated from noisy images, such as low dose computed tomography (CT) scans. In this article, we investigate the possibility of improving the reproducibility of radiomic features calculated on noisy CTs by using generative models for denoising.One traditional denoising method - non-local means - and two generative models - encoder-decoder networks (EDN) and conditional generative adversarial networks (CGANs) - were selected as the test models. We added noise to the sinograms of full dose CTs to mimic low dose CTs with two different levels of noise: low-noise CT and high-noise CT. Models were trained on high-noise CTs and used to denoise low-noise CTs without re-training. We also test the performance of our model in real data, using dataset of same-day repeat low dose CTs to assess the reproducibility of radiomic features in denoised images. The EDN and the CGAN improved the concordance correlation coefficients (CCC) of radiomic features for low-noise images from 0.87 to 0.92 and for high-noise images from 0.68 to 0.92 respectively. Moreover, the EDN and the CGAN improved the test-retest reliability of radiomic features (mean CCC increased from 0.89 to 0.94) based on real low dose CTs. The results show that denoising using EDN and CGANs can improve the reproducibility of radiomic features calculated on noisy CTs. Moreover, images with different noise levels can be denoised to improve the reproducibility using these models without re-training, as long as the noise intensity is equal or lower than that in high-noise CTs. To the authors' knowledge, this is the first effort to improve the reproducibility of radiomic features calculated on low dose CT scans.
Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge
Jul 15, 2017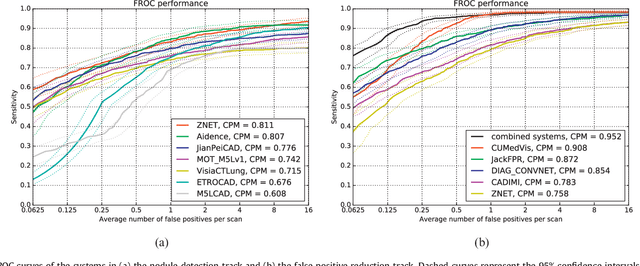
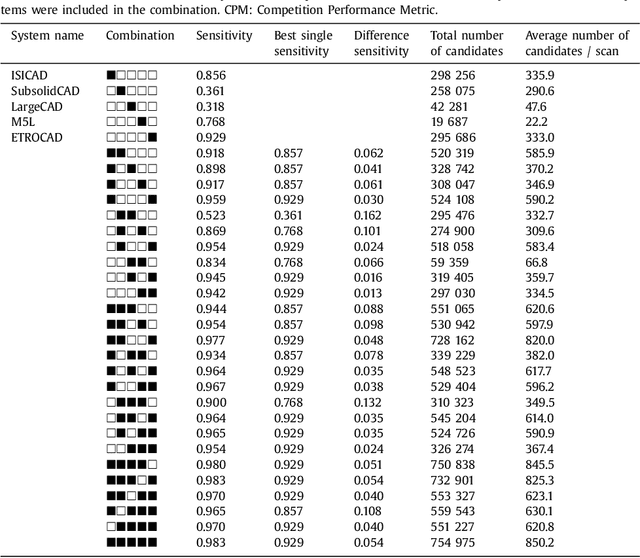
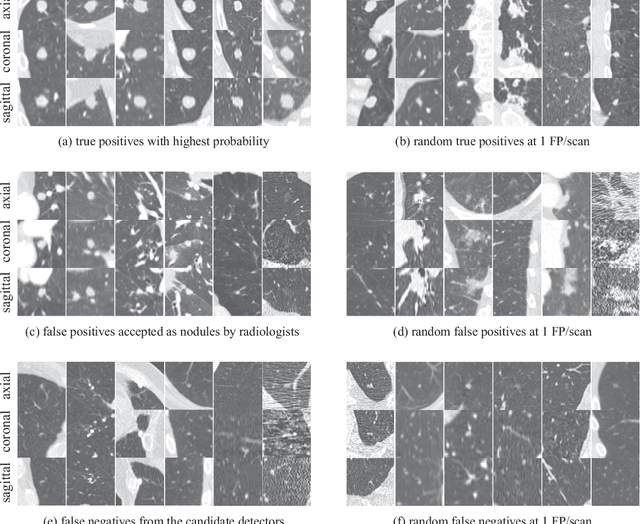
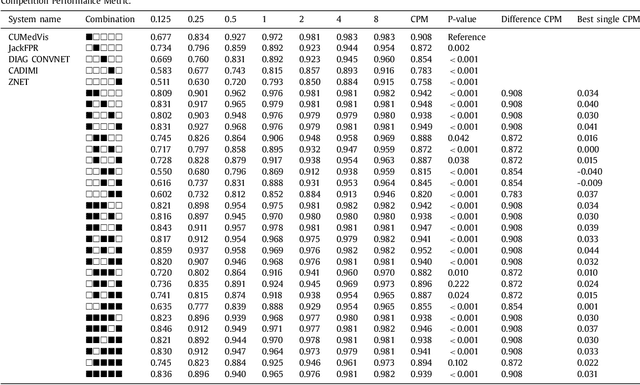
Abstract:Automatic detection of pulmonary nodules in thoracic computed tomography (CT) scans has been an active area of research for the last two decades. However, there have only been few studies that provide a comparative performance evaluation of different systems on a common database. We have therefore set up the LUNA16 challenge, an objective evaluation framework for automatic nodule detection algorithms using the largest publicly available reference database of chest CT scans, the LIDC-IDRI data set. In LUNA16, participants develop their algorithm and upload their predictions on 888 CT scans in one of the two tracks: 1) the complete nodule detection track where a complete CAD system should be developed, or 2) the false positive reduction track where a provided set of nodule candidates should be classified. This paper describes the setup of LUNA16 and presents the results of the challenge so far. Moreover, the impact of combining individual systems on the detection performance was also investigated. It was observed that the leading solutions employed convolutional networks and used the provided set of nodule candidates. The combination of these solutions achieved an excellent sensitivity of over 95% at fewer than 1.0 false positives per scan. This highlights the potential of combining algorithms to improve the detection performance. Our observer study with four expert readers has shown that the best system detects nodules that were missed by expert readers who originally annotated the LIDC-IDRI data. We released this set of additional nodules for further development of CAD systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge