Shahin Heidarian
Spatio-Temporal Hybrid Fusion of CAE and SWIn Transformers for Lung Cancer Malignancy Prediction
Oct 27, 2022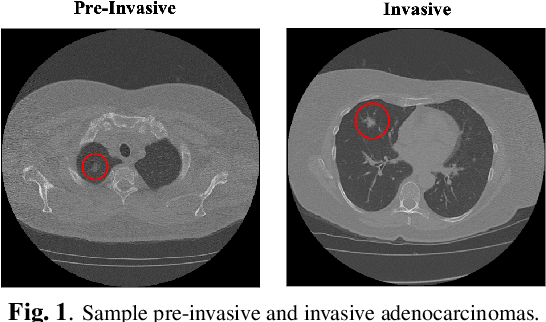
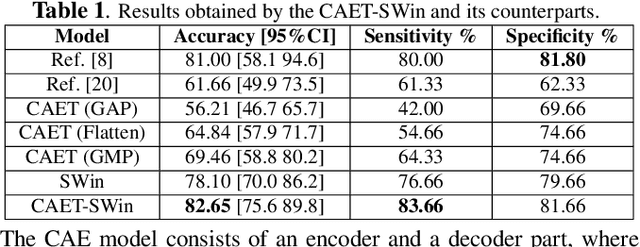
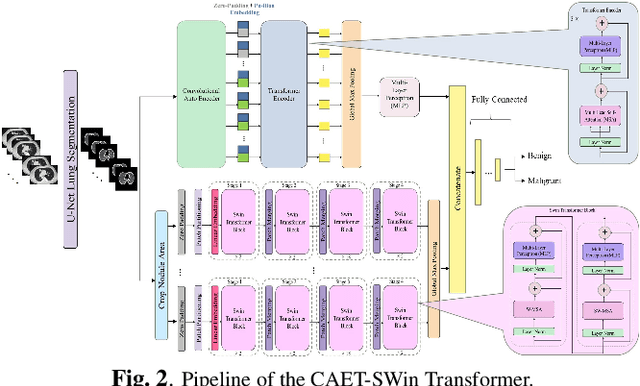
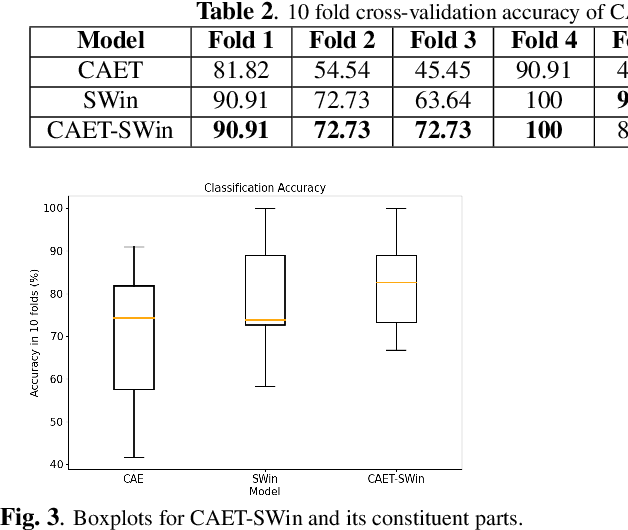
Abstract:The paper proposes a novel hybrid discovery Radiomics framework that simultaneously integrates temporal and spatial features extracted from non-thin chest Computed Tomography (CT) slices to predict Lung Adenocarcinoma (LUAC) malignancy with minimum expert involvement. Lung cancer is the leading cause of mortality from cancer worldwide and has various histologic types, among which LUAC has recently been the most prevalent. LUACs are classified as pre-invasive, minimally invasive, and invasive adenocarcinomas. Timely and accurate knowledge of the lung nodules malignancy leads to a proper treatment plan and reduces the risk of unnecessary or late surgeries. Currently, chest CT scan is the primary imaging modality to assess and predict the invasiveness of LUACs. However, the radiologists' analysis based on CT images is subjective and suffers from a low accuracy compared to the ground truth pathological reviews provided after surgical resections. The proposed hybrid framework, referred to as the CAET-SWin, consists of two parallel paths: (i) The Convolutional Auto-Encoder (CAE) Transformer path that extracts and captures informative features related to inter-slice relations via a modified Transformer architecture, and; (ii) The Shifted Window (SWin) Transformer path, which is a hierarchical vision transformer that extracts nodules' related spatial features from a volumetric CT scan. Extracted temporal (from the CAET-path) and spatial (from the Swin path) are then fused through a fusion path to classify LUACs. Experimental results on our in-house dataset of 114 pathologically proven Sub-Solid Nodules (SSNs) demonstrate that the CAET-SWin significantly improves reliability of the invasiveness prediction task while achieving an accuracy of 82.65%, sensitivity of 83.66%, and specificity of 81.66% using 10-fold cross-validation.
Multi-Content Time-Series Popularity Prediction with Multiple-Model Transformers in MEC Networks
Oct 12, 2022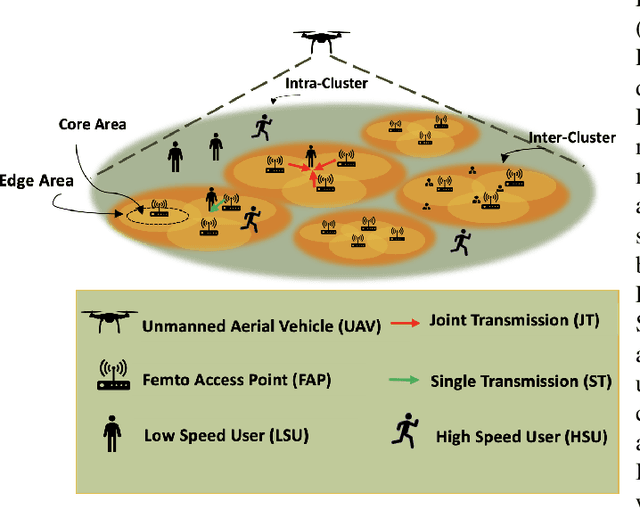
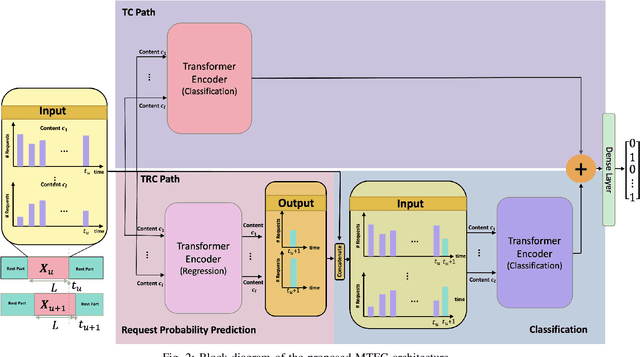
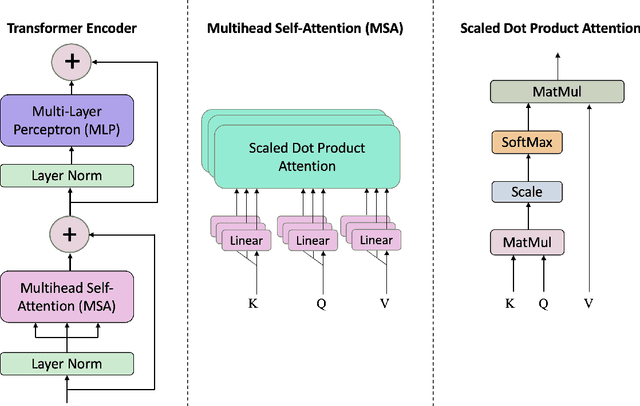
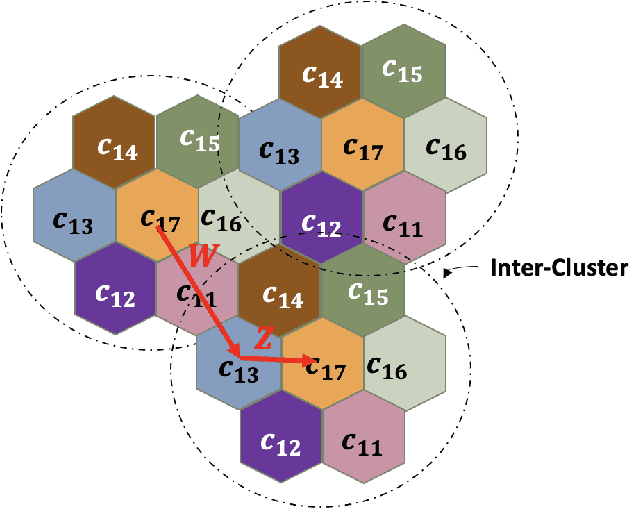
Abstract:Coded/uncoded content placement in Mobile Edge Caching (MEC) has evolved as an efficient solution to meet the significant growth of global mobile data traffic by boosting the content diversity in the storage of caching nodes. To meet the dynamic nature of the historical request pattern of multimedia contents, the main focus of recent researches has been shifted to develop data-driven and real-time caching schemes. In this regard and with the assumption that users' preferences remain unchanged over a short horizon, the Top-K popular contents are identified as the output of the learning model. Most existing datadriven popularity prediction models, however, are not suitable for the coded/uncoded content placement frameworks. On the one hand, in coded/uncoded content placement, in addition to classifying contents into two groups, i.e., popular and nonpopular, the probability of content request is required to identify which content should be stored partially/completely, where this information is not provided by existing data-driven popularity prediction models. On the other hand, the assumption that users' preferences remain unchanged over a short horizon only works for content with a smooth request pattern. To tackle these challenges, we develop a Multiple-model (hybrid) Transformer-based Edge Caching (MTEC) framework with higher generalization ability, suitable for various types of content with different time-varying behavior, that can be adapted with coded/uncoded content placement frameworks. Simulation results corroborate the effectiveness of the proposed MTEC caching framework in comparison to its counterparts in terms of the cache-hit ratio, classification accuracy, and the transferred byte volume.
TEDGE-Caching: Transformer-based Edge Caching Towards 6G Networks
Dec 01, 2021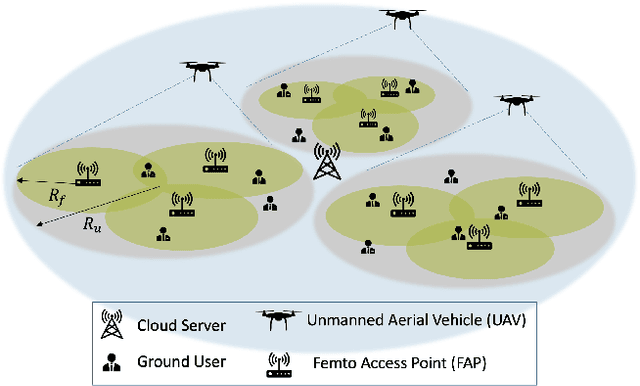

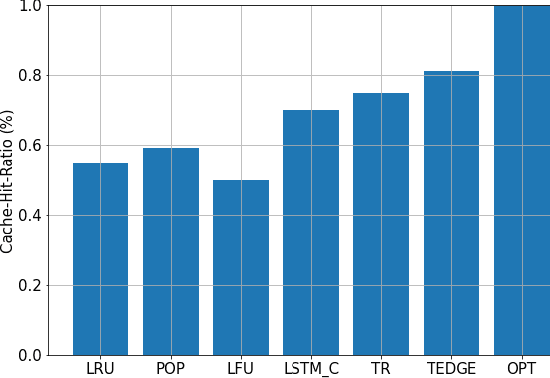
Abstract:As a consequence of the COVID-19 pandemic, the demand for telecommunication for remote learning/working and telemedicine has significantly increased. Mobile Edge Caching (MEC) in the 6G networks has been evolved as an efficient solution to meet the phenomenal growth of the global mobile data traffic by bringing multimedia content closer to the users. Although massive connectivity enabled by MEC networks will significantly increase the quality of communications, there are several key challenges ahead. The limited storage of edge nodes, the large size of multimedia content, and the time-variant users' preferences make it critical to efficiently and dynamically predict the popularity of content to store the most upcoming requested ones before being requested. Recent advancements in Deep Neural Networks (DNNs) have drawn much research attention to predict the content popularity in proactive caching schemes. Existing DNN models in this context, however, suffer from longterm dependencies, computational complexity, and unsuitability for parallel computing. To tackle these challenges, we propose an edge caching framework incorporated with the attention-based Vision Transformer (ViT) neural network, referred to as the Transformer-based Edge (TEDGE) caching, which to the best of our knowledge, is being studied for the first time. Moreover, the TEDGE caching framework requires no data pre-processing and additional contextual information. Simulation results corroborate the effectiveness of the proposed TEDGE caching framework in comparison to its counterparts.
CAE-Transformer: Transformer-based Model to Predict Invasiveness of Lung Adenocarcinoma Subsolid Nodules from Non-thin Section 3D CT Scans
Oct 17, 2021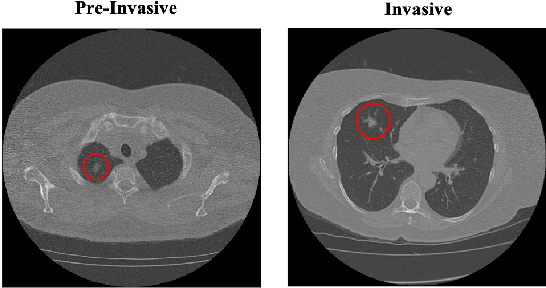
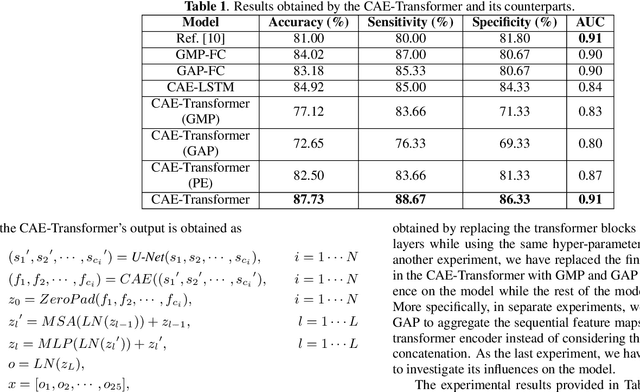
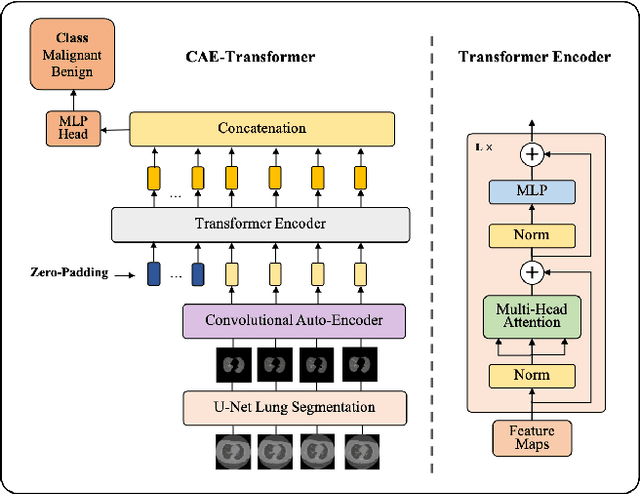
Abstract:Lung cancer is the leading cause of mortality from cancer worldwide and has various histologic types, among which Lung Adenocarcinoma (LAUC) has recently been the most prevalent. Lung adenocarcinomas are classified as pre-invasive, minimally invasive, and invasive adenocarcinomas. Timely and accurate knowledge of the invasiveness of lung nodules leads to a proper treatment plan and reduces the risk of unnecessary or late surgeries. Currently, the primary imaging modality to assess and predict the invasiveness of LAUCs is the chest CT. The results based on CT images, however, are subjective and suffer from a low accuracy compared to the ground truth pathological reviews provided after surgical resections. In this paper, a predictive transformer-based framework, referred to as the "CAE-Transformer", is developed to classify LAUCs. The CAE-Transformer utilizes a Convolutional Auto-Encoder (CAE) to automatically extract informative features from CT slices, which are then fed to a modified transformer model to capture global inter-slice relations. Experimental results on our in-house dataset of 114 pathologically proven Sub-Solid Nodules (SSNs) demonstrate the superiority of the CAE-Transformer over the histogram/radiomics-based models and its deep learning-based counterparts, achieving an accuracy of 87.73%, sensitivity of 88.67%, specificity of 86.33%, and AUC of 0.913, using a 10-fold cross-validation.
Robust Automated Framework for COVID-19 Disease Identification from a Multicenter Dataset of Chest CT Scans
Sep 26, 2021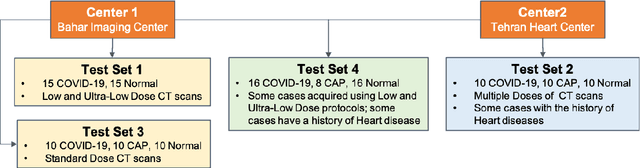
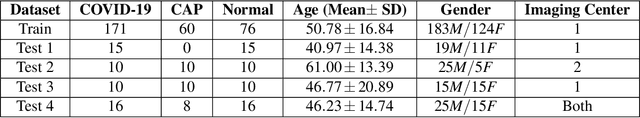
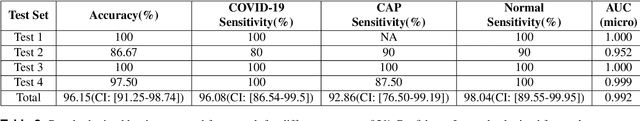
Abstract:The objective of this study is to develop a robust deep learning-based framework to distinguish COVID-19, Community-Acquired Pneumonia (CAP), and Normal cases based on chest CT scans acquired in different imaging centers using various protocols, and radiation doses. We showed that while our proposed model is trained on a relatively small dataset acquired from only one imaging center using a specific scanning protocol, the model performs well on heterogeneous test sets obtained by multiple scanners using different technical parameters. We also showed that the model can be updated via an unsupervised approach to cope with the data shift between the train and test sets and enhance the robustness of the model upon receiving a new external dataset from a different center. We adopted an ensemble architecture to aggregate the predictions from multiple versions of the model. For initial training and development purposes, an in-house dataset of 171 COVID-19, 60 CAP, and 76 Normal cases was used, which contained volumetric CT scans acquired from one imaging center using a constant standard radiation dose scanning protocol. To evaluate the model, we collected four different test sets retrospectively to investigate the effects of the shifts in the data characteristics on the model's performance. Among the test cases, there were CT scans with similar characteristics as the train set as well as noisy low-dose and ultra-low dose CT scans. In addition, some test CT scans were obtained from patients with a history of cardiovascular diseases or surgeries. The entire test dataset used in this study contained 51 COVID-19, 28 CAP, and 51 Normal cases. Experimental results indicate that our proposed framework performs well on all test sets achieving total accuracy of 96.15% (95%CI: [91.25-98.74]), COVID-19 sensitivity of 96.08% (95%CI: [86.54-99.5]), CAP sensitivity of 92.86% (95%CI: [76.50-99.19]).
COVID-Rate: An Automated Framework for Segmentation of COVID-19 Lesions from Chest CT Scans
Jul 04, 2021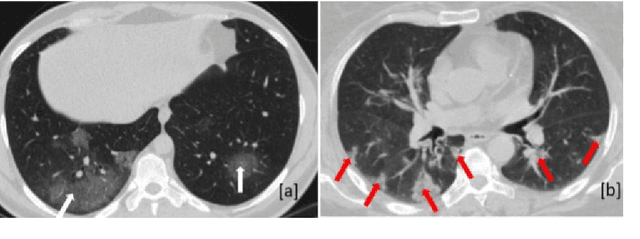

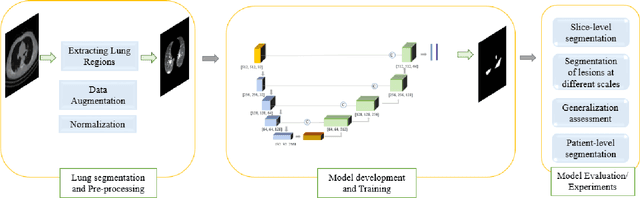
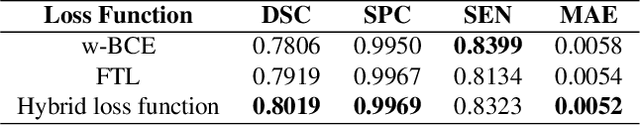
Abstract:Novel Coronavirus disease (COVID-19) is a highly contagious respiratory infection that has had devastating effects on the world. Recently, new COVID-19 variants are emerging making the situation more challenging and threatening. Evaluation and quantification of COVID-19 lung abnormalities based on chest Computed Tomography (CT) scans can help determining the disease stage, efficiently allocating limited healthcare resources, and making informed treatment decisions. During pandemic era, however, visual assessment and quantification of COVID-19 lung lesions by expert radiologists become expensive and prone to error, which raises an urgent quest to develop practical autonomous solutions. In this context, first, the paper introduces an open access COVID-19 CT segmentation dataset containing 433 CT images from 82 patients that have been annotated by an expert radiologist. Second, a Deep Neural Network (DNN)-based framework is proposed, referred to as the COVID-Rate, that autonomously segments lung abnormalities associated with COVID-19 from chest CT scans. Performance of the proposed COVID-Rate framework is evaluated through several experiments based on the introduced and external datasets. The results show a dice score of 0:802 and specificity and sensitivity of 0:997 and 0:832, respectively. Furthermore, the results indicate that the COVID-Rate model can efficiently segment COVID-19 lesions in both 2D CT images and whole lung volumes. Results on the external dataset illustrate generalization capabilities of the COVID-Rate model to CT images obtained from a different scanner.
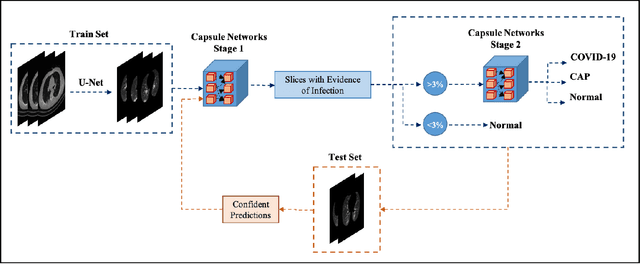
Human-level COVID-19 Diagnosis from Low-dose CT Scans Using a Two-stage Time-distributed Capsule Network
May 31, 2021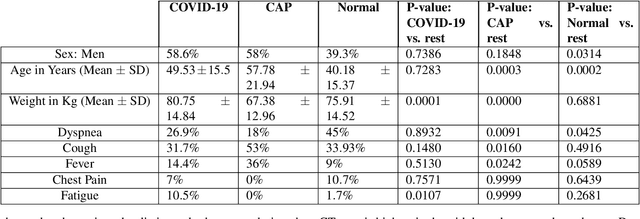
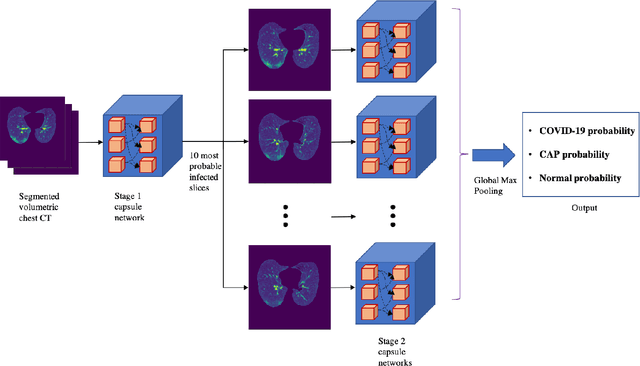
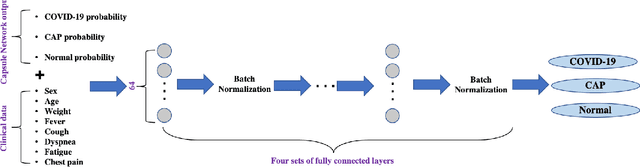
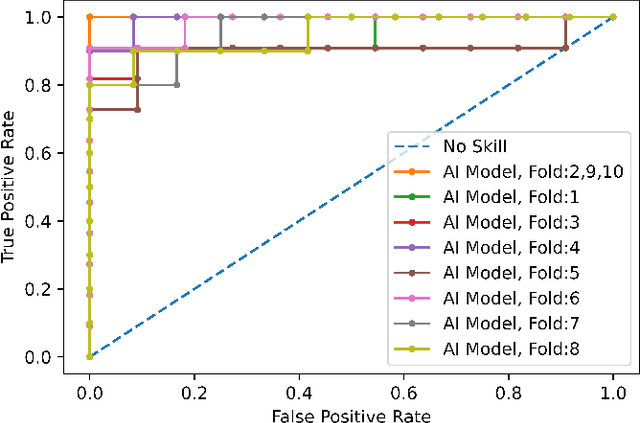
Abstract:Reverse transcription-polymerase chain reaction (RT-PCR) is currently the gold standard in COVID-19 diagnosis. It can, however, take days to provide the diagnosis, and false negative rate is relatively high. Imaging, in particular chest computed tomography (CT), can assist with diagnosis and assessment of this disease. Nevertheless, it is shown that standard dose CT scan gives significant radiation burden to patients, especially those in need of multiple scans. In this study, we consider low-dose and ultra-low-dose (LDCT and ULDCT) scan protocols that reduce the radiation exposure close to that of a single X-Ray, while maintaining an acceptable resolution for diagnosis purposes. Since thoracic radiology expertise may not be widely available during the pandemic, we develop an Artificial Intelligence (AI)-based framework using a collected dataset of LDCT/ULDCT scans, to study the hypothesis that the AI model can provide human-level performance. The AI model uses a two stage capsule network architecture and can rapidly classify COVID-19, community acquired pneumonia (CAP), and normal cases, using LDCT/ULDCT scans. The AI model achieves COVID-19 sensitivity of 89.5% +\- 0.11, CAP sensitivity of 95% +\- 0.11, normal cases sensitivity (specificity) of 85.7% +\- 0.16, and accuracy of 90% +\- 0.06. By incorporating clinical data (demographic and symptoms), the performance further improves to COVID-19 sensitivity of 94.3% +\- pm 0.05, CAP sensitivity of 96.7% +\- 0.07, normal cases sensitivity (specificity) of 91% +\- 0.09 , and accuracy of 94.1% +\- 0.03. The proposed AI model achieves human-level diagnosis based on the LDCT/ULDCT scans with reduced radiation exposure. We believe that the proposed AI model has the potential to assist the radiologists to accurately and promptly diagnose COVID-19 infection and help control the transmission chain during the pandemic.
CT-CAPS: Feature Extraction-based Automated Framework for COVID-19 Disease Identification from Chest CT Scans using Capsule Networks
Oct 30, 2020
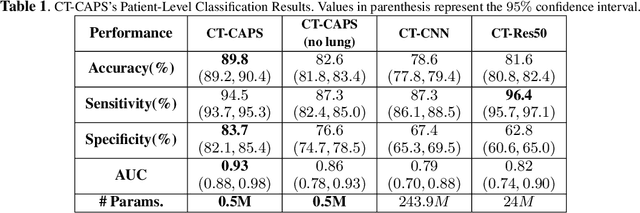
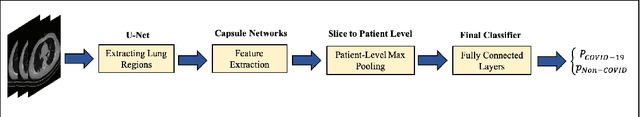
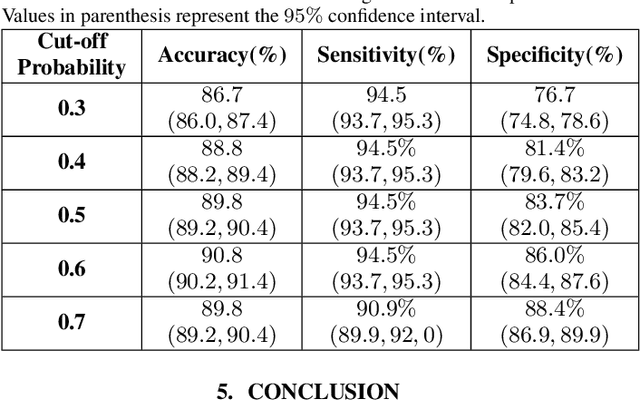
Abstract:The global outbreak of the novel corona virus (COVID-19) disease has drastically impacted the world and led to one of the most challenging crisis across the globe since World War II. The early diagnosis and isolation of COVID-19 positive cases are considered as crucial steps towards preventing the spread of the disease and flattening the epidemic curve. Chest Computed Tomography (CT) scan is a highly sensitive, rapid, and accurate diagnostic technique that can complement Reverse Transcription Polymerase Chain Reaction (RT-PCR) test. Recently, deep learning-based models, mostly based on Convolutional Neural Networks (CNN), have shown promising diagnostic results. CNNs, however, are incapable of capturing spatial relations between image instances and require large datasets. Capsule Networks, on the other hand, can capture spatial relations, require smaller datasets, and have considerably fewer parameters. In this paper, a Capsule network framework, referred to as the "CT-CAPS", is presented to automatically extract distinctive features of chest CT scans. These features, which are extracted from the layer before the final capsule layer, are then leveraged to differentiate COVID-19 from Non-COVID cases. The experiments on our in-house dataset of 307 patients show the state-of-the-art performance with the accuracy of 90.8%, sensitivity of 94.5%, and specificity of 86.0%.
COVID-FACT: A Fully-Automated Capsule Network-based Framework for Identification of COVID-19 Cases from Chest CT scans
Oct 30, 2020
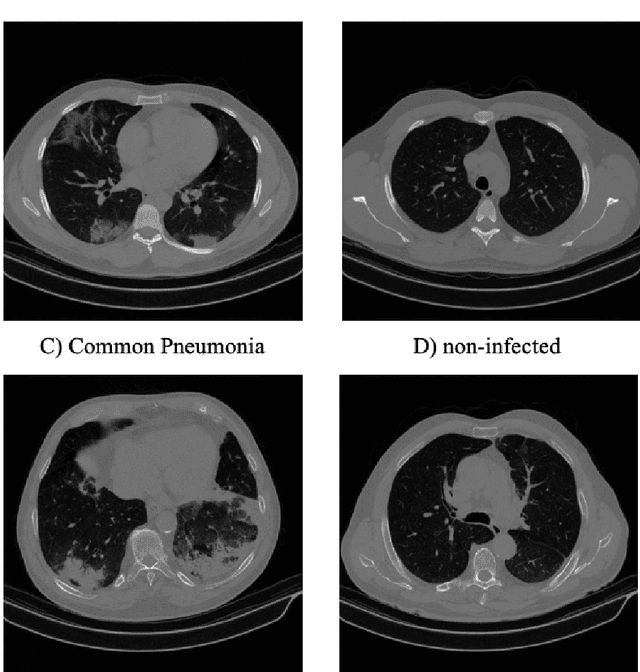
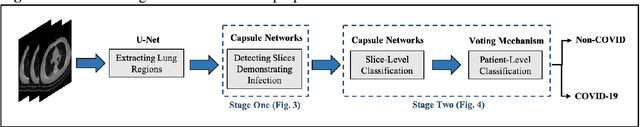
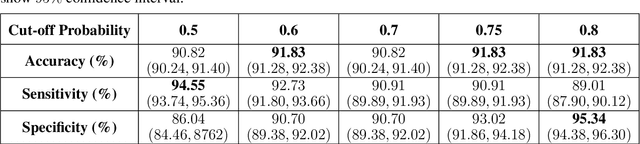
Abstract:The newly discovered Corona virus Disease 2019 (COVID-19) has been globally spreading and causing hundreds of thousands of deaths around the world as of its first emergence in late 2019. Computed tomography (CT) scans have shown distinctive features and higher sensitivity compared to other diagnostic tests, in particular the current gold standard, i.e., the Reverse Transcription Polymerase Chain Reaction (RT-PCR) test. Current deep learning-based algorithms are mainly developed based on Convolutional Neural Networks (CNNs) to identify COVID-19 pneumonia cases. CNNs, however, require extensive data augmentation and large datasets to identify detailed spatial relations between image instances. Furthermore, existing algorithms utilizing CT scans, either extend slice-level predictions to patient-level ones using a simple thresholding mechanism or rely on a sophisticated infection segmentation to identify the disease. In this paper, we propose a two-stage fully-automated CT-based framework for identification of COVID-19 positive cases referred to as the "COVID-FACT". COVID-FACT utilizes Capsule Networks, as its main building blocks and is, therefore, capable of capturing spatial information. In particular, to make the proposed COVID-FACT independent from sophisticated segmentation of the area of infection, slices demonstrating infection are detected at the first stage and the second stage is responsible for classifying patients into COVID and non-COVID cases. COVID-FACT detects slices with infection, and identifies positive COVID-19 cases using an in-house CT scan dataset, containing COVID-19, community acquired pneumonia, and normal cases. Based on our experiments, COVID-FACT achieves an accuracy of 90.82%, a sensitivity of 94.55%, a specificity of 86.04%, and an Area Under the Curve (AUC) of 0.98, while depending on far less supervision and annotation, in comparison to its counterparts.
COVID-CT-MD: COVID-19 Computed Tomography Scan Dataset Applicable in Machine Learning and Deep Learning
Sep 28, 2020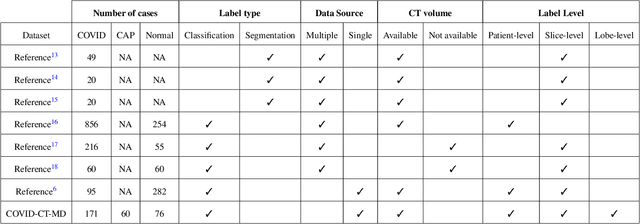
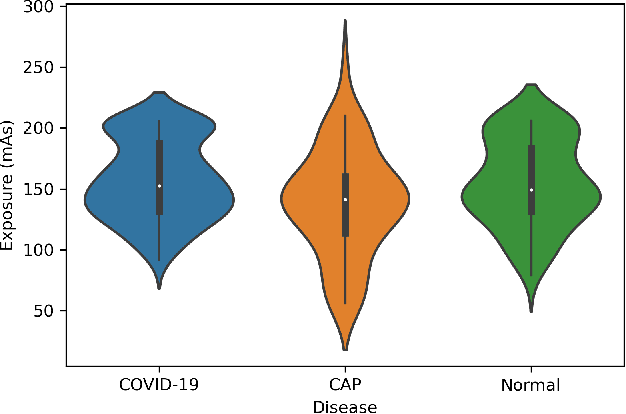

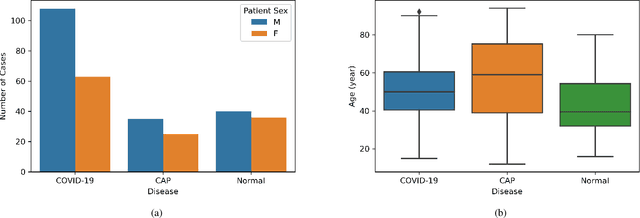
Abstract:Novel Coronavirus (COVID-19) has drastically overwhelmed more than 200 countries affecting millions and claiming almost 1 million lives, since its emergence in late 2019. This highly contagious disease can easily spread, and if not controlled in a timely fashion, can rapidly incapacitate healthcare systems. The current standard diagnosis method, the Reverse Transcription Polymerase Chain Reaction (RT- PCR), is time consuming, and subject to low sensitivity. Chest Radiograph (CXR), the first imaging modality to be used, is readily available and gives immediate results. However, it has notoriously lower sensitivity than Computed Tomography (CT), which can be used efficiently to complement other diagnostic methods. This paper introduces a new COVID-19 CT scan dataset, referred to as COVID-CT-MD, consisting of not only COVID-19 cases, but also healthy and subjects infected by Community Acquired Pneumonia (CAP). COVID-CT-MD dataset, which is accompanied with lobe-level, slice-level and patient-level labels, has the potential to facilitate the COVID-19 research, in particular COVID-CT-MD can assist in development of advanced Machine Learning (ML) and Deep Neural Network (DNN) based solutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge