Moezedin Javad Rafiee
Data Shapley Value for Handling Noisy Labels: An application in Screening COVID-19 Pneumonia from Chest CT Scans
Oct 17, 2021
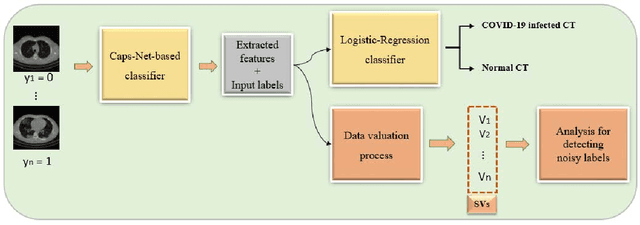
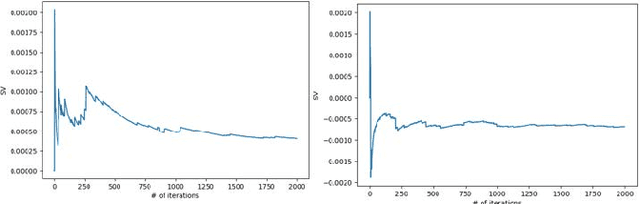
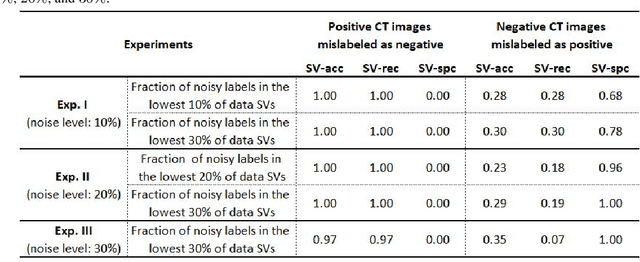
Abstract:A long-standing challenge of deep learning models involves how to handle noisy labels, especially in applications where human lives are at stake. Adoption of the data Shapley Value (SV), a cooperative game theoretical approach, is an intelligent valuation solution to tackle the issue of noisy labels. Data SV can be used together with a learning model and an evaluation metric to validate each training point's contribution to the model's performance. The SV of a data point, however, is not unique and depends on the learning model, the evaluation metric, and other data points collaborating in the training game. However, effects of utilizing different evaluation metrics for computation of the SV, detecting the noisy labels, and measuring the data points' importance has not yet been thoroughly investigated. In this context, we performed a series of comparative analyses to assess SV's capabilities to detect noisy input labels when measured by different evaluation metrics. Our experiments on COVID-19-infected of CT images illustrate that although the data SV can effectively identify noisy labels, adoption of different evaluation metric can significantly influence its ability to identify noisy labels from different data classes. Specifically, we demonstrate that the SV greatly depends on the associated evaluation metric.
Robust Automated Framework for COVID-19 Disease Identification from a Multicenter Dataset of Chest CT Scans
Sep 26, 2021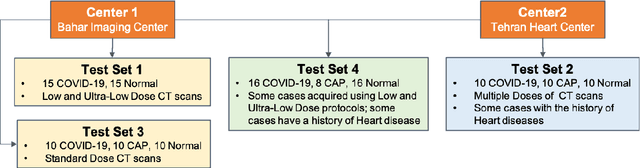
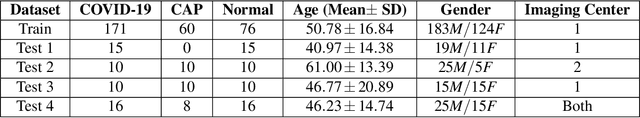
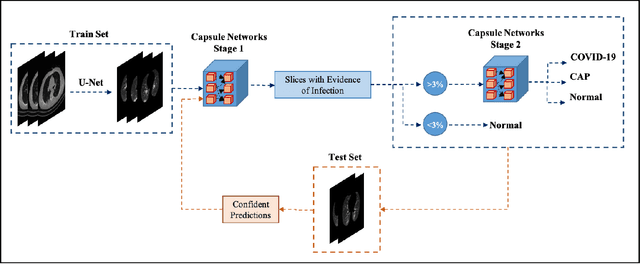
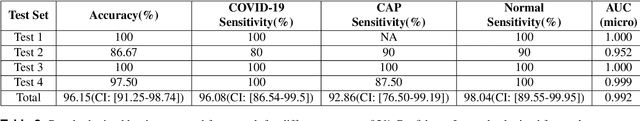
Abstract:The objective of this study is to develop a robust deep learning-based framework to distinguish COVID-19, Community-Acquired Pneumonia (CAP), and Normal cases based on chest CT scans acquired in different imaging centers using various protocols, and radiation doses. We showed that while our proposed model is trained on a relatively small dataset acquired from only one imaging center using a specific scanning protocol, the model performs well on heterogeneous test sets obtained by multiple scanners using different technical parameters. We also showed that the model can be updated via an unsupervised approach to cope with the data shift between the train and test sets and enhance the robustness of the model upon receiving a new external dataset from a different center. We adopted an ensemble architecture to aggregate the predictions from multiple versions of the model. For initial training and development purposes, an in-house dataset of 171 COVID-19, 60 CAP, and 76 Normal cases was used, which contained volumetric CT scans acquired from one imaging center using a constant standard radiation dose scanning protocol. To evaluate the model, we collected four different test sets retrospectively to investigate the effects of the shifts in the data characteristics on the model's performance. Among the test cases, there were CT scans with similar characteristics as the train set as well as noisy low-dose and ultra-low dose CT scans. In addition, some test CT scans were obtained from patients with a history of cardiovascular diseases or surgeries. The entire test dataset used in this study contained 51 COVID-19, 28 CAP, and 51 Normal cases. Experimental results indicate that our proposed framework performs well on all test sets achieving total accuracy of 96.15% (95%CI: [91.25-98.74]), COVID-19 sensitivity of 96.08% (95%CI: [86.54-99.5]), CAP sensitivity of 92.86% (95%CI: [76.50-99.19]).
COVID-Rate: An Automated Framework for Segmentation of COVID-19 Lesions from Chest CT Scans
Jul 04, 2021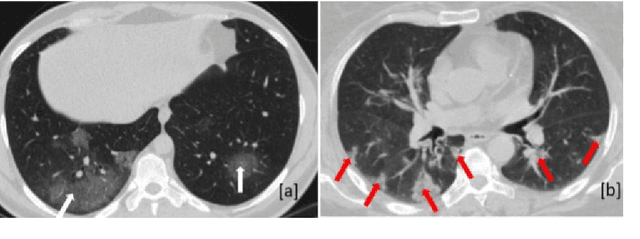

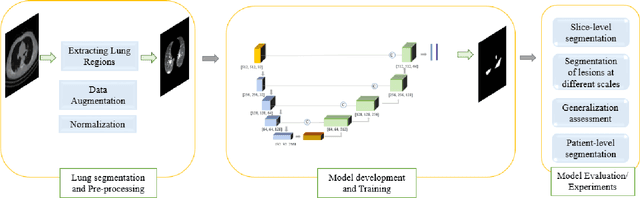
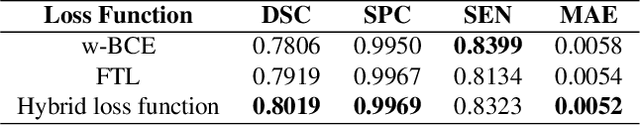
Abstract:Novel Coronavirus disease (COVID-19) is a highly contagious respiratory infection that has had devastating effects on the world. Recently, new COVID-19 variants are emerging making the situation more challenging and threatening. Evaluation and quantification of COVID-19 lung abnormalities based on chest Computed Tomography (CT) scans can help determining the disease stage, efficiently allocating limited healthcare resources, and making informed treatment decisions. During pandemic era, however, visual assessment and quantification of COVID-19 lung lesions by expert radiologists become expensive and prone to error, which raises an urgent quest to develop practical autonomous solutions. In this context, first, the paper introduces an open access COVID-19 CT segmentation dataset containing 433 CT images from 82 patients that have been annotated by an expert radiologist. Second, a Deep Neural Network (DNN)-based framework is proposed, referred to as the COVID-Rate, that autonomously segments lung abnormalities associated with COVID-19 from chest CT scans. Performance of the proposed COVID-Rate framework is evaluated through several experiments based on the introduced and external datasets. The results show a dice score of 0:802 and specificity and sensitivity of 0:997 and 0:832, respectively. Furthermore, the results indicate that the COVID-Rate model can efficiently segment COVID-19 lesions in both 2D CT images and whole lung volumes. Results on the external dataset illustrate generalization capabilities of the COVID-Rate model to CT images obtained from a different scanner.
Human-level COVID-19 Diagnosis from Low-dose CT Scans Using a Two-stage Time-distributed Capsule Network
May 31, 2021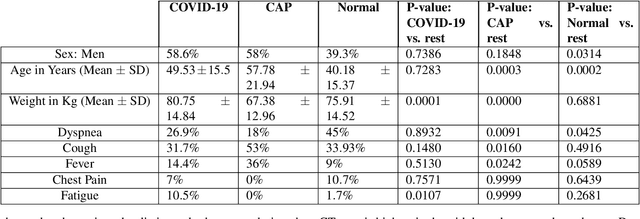
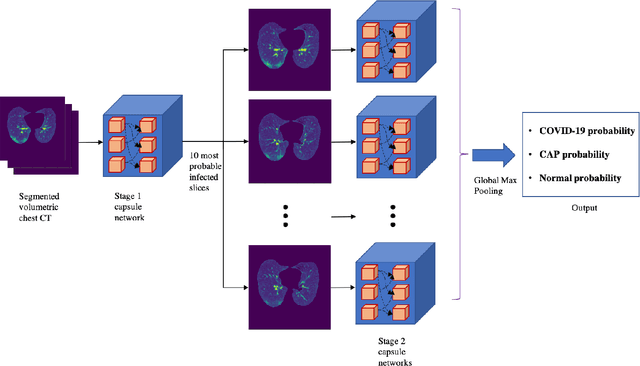
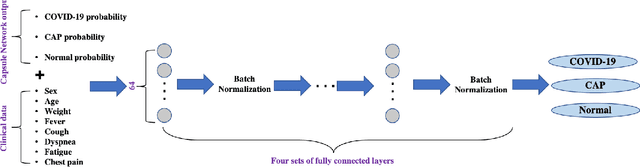
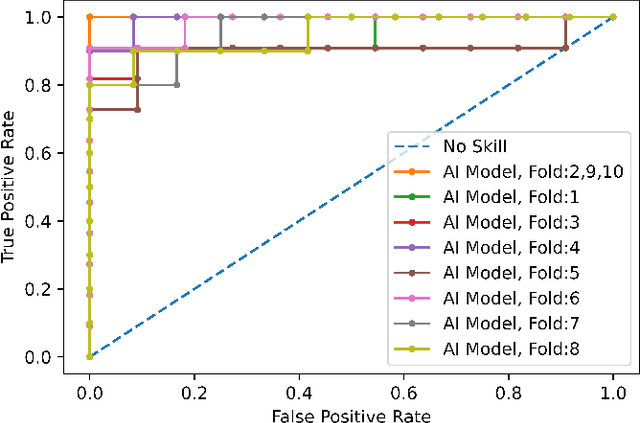
Abstract:Reverse transcription-polymerase chain reaction (RT-PCR) is currently the gold standard in COVID-19 diagnosis. It can, however, take days to provide the diagnosis, and false negative rate is relatively high. Imaging, in particular chest computed tomography (CT), can assist with diagnosis and assessment of this disease. Nevertheless, it is shown that standard dose CT scan gives significant radiation burden to patients, especially those in need of multiple scans. In this study, we consider low-dose and ultra-low-dose (LDCT and ULDCT) scan protocols that reduce the radiation exposure close to that of a single X-Ray, while maintaining an acceptable resolution for diagnosis purposes. Since thoracic radiology expertise may not be widely available during the pandemic, we develop an Artificial Intelligence (AI)-based framework using a collected dataset of LDCT/ULDCT scans, to study the hypothesis that the AI model can provide human-level performance. The AI model uses a two stage capsule network architecture and can rapidly classify COVID-19, community acquired pneumonia (CAP), and normal cases, using LDCT/ULDCT scans. The AI model achieves COVID-19 sensitivity of 89.5% +\- 0.11, CAP sensitivity of 95% +\- 0.11, normal cases sensitivity (specificity) of 85.7% +\- 0.16, and accuracy of 90% +\- 0.06. By incorporating clinical data (demographic and symptoms), the performance further improves to COVID-19 sensitivity of 94.3% +\- pm 0.05, CAP sensitivity of 96.7% +\- 0.07, normal cases sensitivity (specificity) of 91% +\- 0.09 , and accuracy of 94.1% +\- 0.03. The proposed AI model achieves human-level diagnosis based on the LDCT/ULDCT scans with reduced radiation exposure. We believe that the proposed AI model has the potential to assist the radiologists to accurately and promptly diagnose COVID-19 infection and help control the transmission chain during the pandemic.
Diagnosis/Prognosis of COVID-19 Images: Challenges, Opportunities, and Applications
Dec 28, 2020
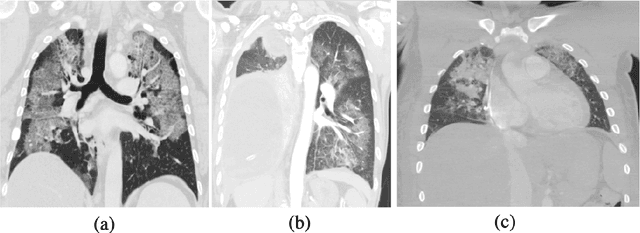
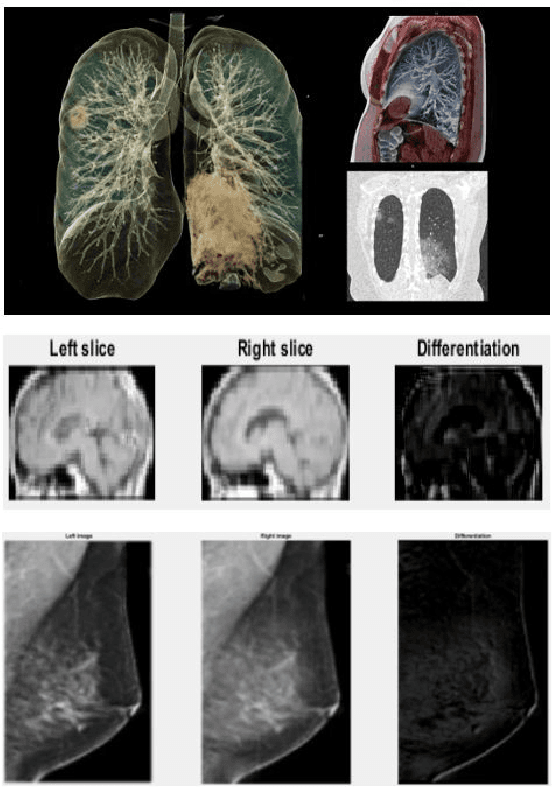
Abstract:The novel Coronavirus disease, COVID-19, has rapidly and abruptly changed the world as we knew in 2020. It becomes the most unprecedent challenge to analytic epidemiology in general and signal processing theories in specific. Given its high contingency nature and adverse effects across the world, it is important to develop efficient processing/learning models to overcome this pandemic and be prepared for potential future ones. In this regard, medical imaging plays an important role for the management of COVID-19. Human-centered interpretation of medical images is, however, tedious and can be subjective. This has resulted in a surge of interest to develop Radiomics models for analysis and interpretation of medical images. Signal Processing (SP) and Deep Learning (DL) models can assist in development of robust Radiomics solutions for diagnosis/prognosis, severity assessment, treatment response, and monitoring of COVID-19 patients. In this article, we aim to present an overview of the current state, challenges, and opportunities of developing SP/DL-empowered models for diagnosis (screening/monitoring) and prognosis (outcome prediction and severity assessment) of COVID-19 infection. More specifically, the article starts by elaborating the latest development on the theoretical framework of analytic epidemiology and hypersignal processing for COVID-19. Afterwards, imaging modalities and Radiological characteristics of COVID-19 are discussed. SL/DL-based Radiomic models specific to the analysis of COVID-19 infection are then described covering the following four domains: Segmentation of COVID-19 lesions; Predictive models for outcome prediction; Severity assessment, and; Diagnosis/classification models. Finally, open problems and opportunities are presented in detail.
CT-CAPS: Feature Extraction-based Automated Framework for COVID-19 Disease Identification from Chest CT Scans using Capsule Networks
Oct 30, 2020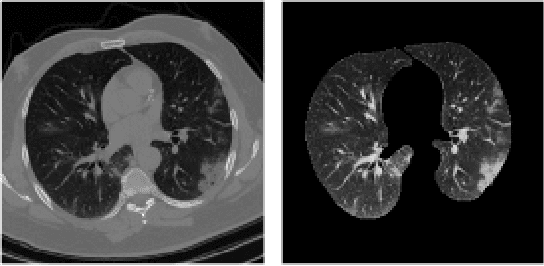
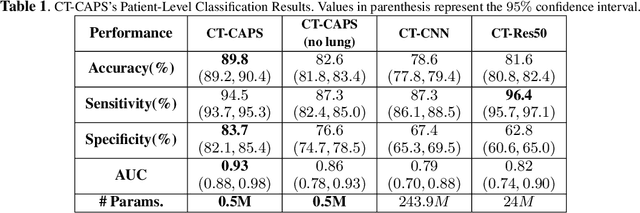
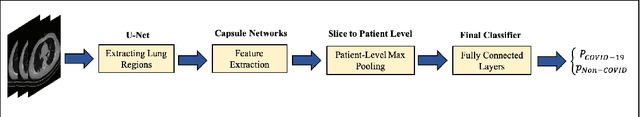
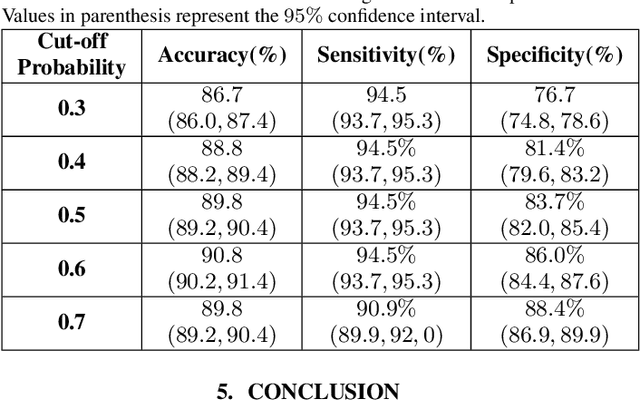
Abstract:The global outbreak of the novel corona virus (COVID-19) disease has drastically impacted the world and led to one of the most challenging crisis across the globe since World War II. The early diagnosis and isolation of COVID-19 positive cases are considered as crucial steps towards preventing the spread of the disease and flattening the epidemic curve. Chest Computed Tomography (CT) scan is a highly sensitive, rapid, and accurate diagnostic technique that can complement Reverse Transcription Polymerase Chain Reaction (RT-PCR) test. Recently, deep learning-based models, mostly based on Convolutional Neural Networks (CNN), have shown promising diagnostic results. CNNs, however, are incapable of capturing spatial relations between image instances and require large datasets. Capsule Networks, on the other hand, can capture spatial relations, require smaller datasets, and have considerably fewer parameters. In this paper, a Capsule network framework, referred to as the "CT-CAPS", is presented to automatically extract distinctive features of chest CT scans. These features, which are extracted from the layer before the final capsule layer, are then leveraged to differentiate COVID-19 from Non-COVID cases. The experiments on our in-house dataset of 307 patients show the state-of-the-art performance with the accuracy of 90.8%, sensitivity of 94.5%, and specificity of 86.0%.
COVID-FACT: A Fully-Automated Capsule Network-based Framework for Identification of COVID-19 Cases from Chest CT scans
Oct 30, 2020
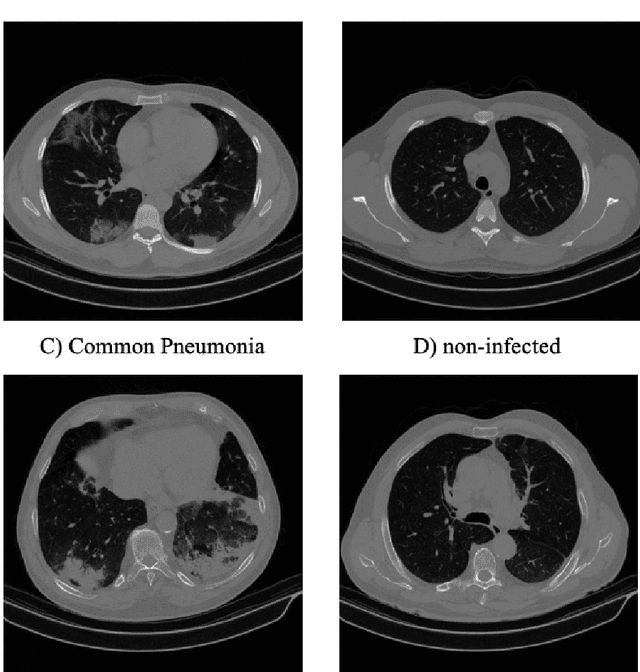
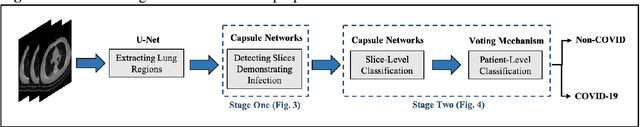
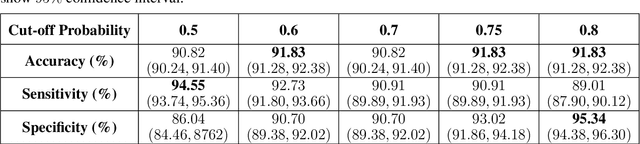
Abstract:The newly discovered Corona virus Disease 2019 (COVID-19) has been globally spreading and causing hundreds of thousands of deaths around the world as of its first emergence in late 2019. Computed tomography (CT) scans have shown distinctive features and higher sensitivity compared to other diagnostic tests, in particular the current gold standard, i.e., the Reverse Transcription Polymerase Chain Reaction (RT-PCR) test. Current deep learning-based algorithms are mainly developed based on Convolutional Neural Networks (CNNs) to identify COVID-19 pneumonia cases. CNNs, however, require extensive data augmentation and large datasets to identify detailed spatial relations between image instances. Furthermore, existing algorithms utilizing CT scans, either extend slice-level predictions to patient-level ones using a simple thresholding mechanism or rely on a sophisticated infection segmentation to identify the disease. In this paper, we propose a two-stage fully-automated CT-based framework for identification of COVID-19 positive cases referred to as the "COVID-FACT". COVID-FACT utilizes Capsule Networks, as its main building blocks and is, therefore, capable of capturing spatial information. In particular, to make the proposed COVID-FACT independent from sophisticated segmentation of the area of infection, slices demonstrating infection are detected at the first stage and the second stage is responsible for classifying patients into COVID and non-COVID cases. COVID-FACT detects slices with infection, and identifies positive COVID-19 cases using an in-house CT scan dataset, containing COVID-19, community acquired pneumonia, and normal cases. Based on our experiments, COVID-FACT achieves an accuracy of 90.82%, a sensitivity of 94.55%, a specificity of 86.04%, and an Area Under the Curve (AUC) of 0.98, while depending on far less supervision and annotation, in comparison to its counterparts.
COVID-CT-MD: COVID-19 Computed Tomography Scan Dataset Applicable in Machine Learning and Deep Learning
Sep 28, 2020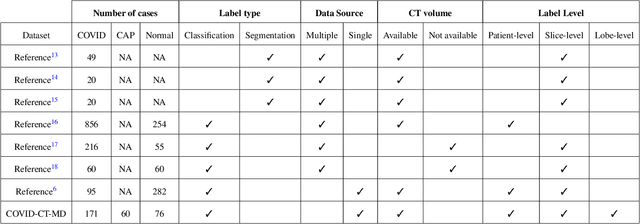
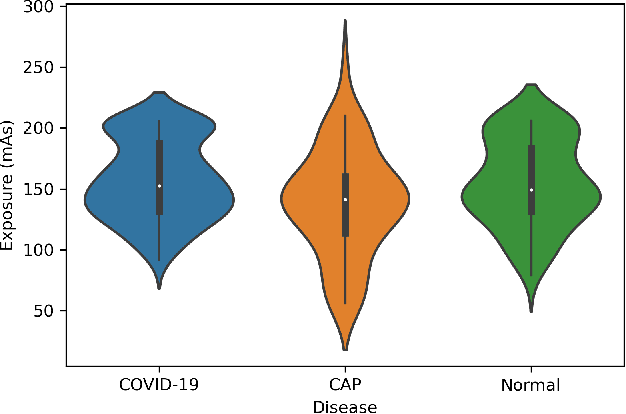

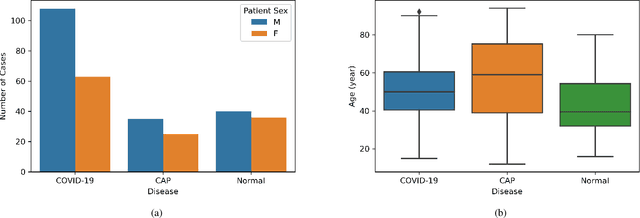
Abstract:Novel Coronavirus (COVID-19) has drastically overwhelmed more than 200 countries affecting millions and claiming almost 1 million lives, since its emergence in late 2019. This highly contagious disease can easily spread, and if not controlled in a timely fashion, can rapidly incapacitate healthcare systems. The current standard diagnosis method, the Reverse Transcription Polymerase Chain Reaction (RT- PCR), is time consuming, and subject to low sensitivity. Chest Radiograph (CXR), the first imaging modality to be used, is readily available and gives immediate results. However, it has notoriously lower sensitivity than Computed Tomography (CT), which can be used efficiently to complement other diagnostic methods. This paper introduces a new COVID-19 CT scan dataset, referred to as COVID-CT-MD, consisting of not only COVID-19 cases, but also healthy and subjects infected by Community Acquired Pneumonia (CAP). COVID-CT-MD dataset, which is accompanied with lobe-level, slice-level and patient-level labels, has the potential to facilitate the COVID-19 research, in particular COVID-CT-MD can assist in development of advanced Machine Learning (ML) and Deep Neural Network (DNN) based solutions.
MIXCAPS: A Capsule Network-based Mixture of Experts for Lung Nodule Malignancy Prediction
Aug 13, 2020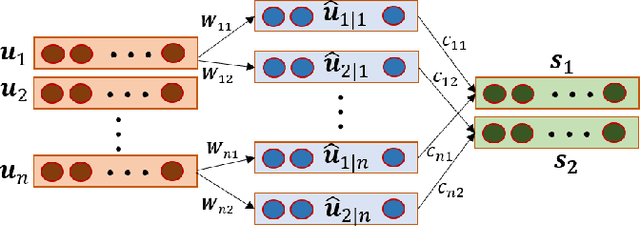
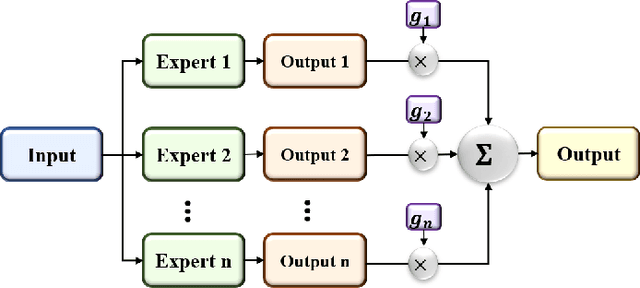
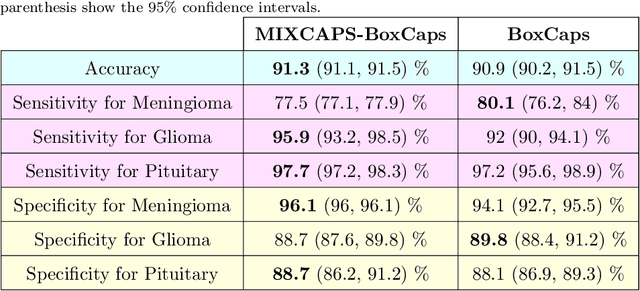
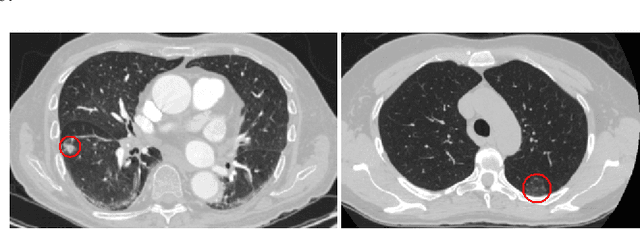
Abstract:Lung diseases including infections such as Pneumonia, Tuberculosis, and novel Coronavirus (COVID-19), together with Lung Cancer are significantly widespread and are, typically, considered life threatening. In particular, lung cancer is among the most common and deadliest cancers with a low 5-year survival rate. Timely diagnosis of lung cancer is, therefore, of paramount importance as it can save countless lives. In this regard, deep learning radiomics solutions have the promise of extracting the most useful features on their own in an end-to-end fashion without having access to the annotated boundaries. Among different deep learning models, Capsule Networks are proposed to overcome shortcomings of the Convolutional Neural Networks (CNN) such as their inability to recognize detailed spatial relations. Capsule networks have so far shown satisfying performance in medical imaging problems. Capitalizing on their success, in this study, we propose a novel capsule network-based mixture of experts, referred to as the MIXCAPS. The proposed MIXCAPS architecture takes advantage of not only the capsule network's capabilities to handle small datasets, but also automatically splitting dataset through a convolutional gating network. MIXCAPS enables capsule network experts to specialize on different subsets of the data. Our results show that MIXCAPS outperforms a single capsule network and a mixture of CNNs, with an accuracy of 92.88%, sensitivity of 93.2%, specificity of 92.3% and area under the curve of 0.963. Our experiments also show that there is a relation between the gate outputs and a couple of hand-crafted features, illustrating explainable nature of the proposed MIXCAPS. To further evaluate generalization capabilities of the proposed MIXCAPS architecture, additional experiments on a brain tumor dataset are performed showing potentials of MIXCAPS for detection of tumors related to other organs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge