Đorđe Miladinović
Learning to Drop Out: An Adversarial Approach to Training Sequence VAEs
Sep 26, 2022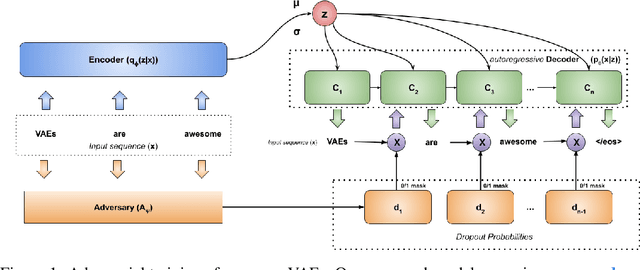
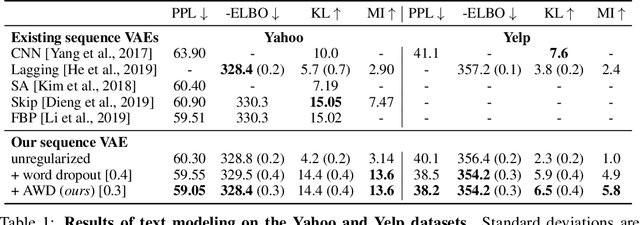
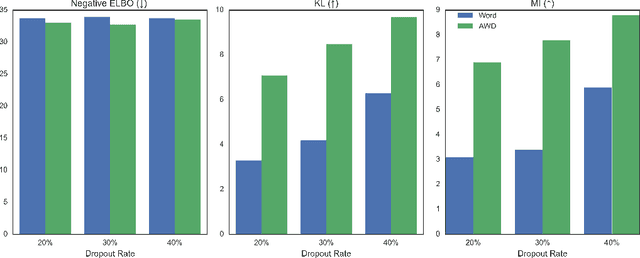
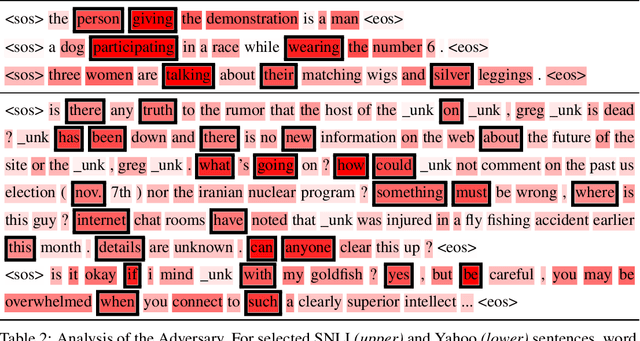
Abstract:In principle, applying variational autoencoders (VAEs) to sequential data offers a method for controlled sequence generation, manipulation, and structured representation learning. However, training sequence VAEs is challenging: autoregressive decoders can often explain the data without utilizing the latent space, known as posterior collapse. To mitigate this, state-of-the-art models weaken the powerful decoder by applying uniformly random dropout to the decoder input. We show theoretically that this removes pointwise mutual information provided by the decoder input, which is compensated for by utilizing the latent space. We then propose an adversarial training strategy to achieve information-based stochastic dropout. Compared to uniform dropout on standard text benchmark datasets, our targeted approach increases both sequence modeling performance and the information captured in the latent space.
Spatially Dependent U-Nets: Highly Accurate Architectures for Medical Imaging Segmentation
Mar 22, 2021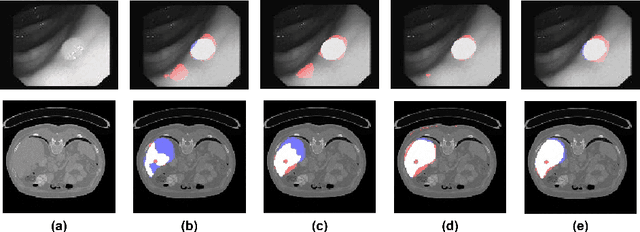
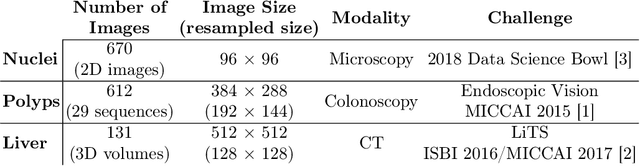
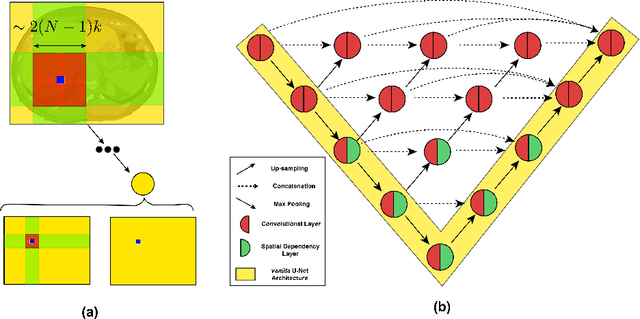
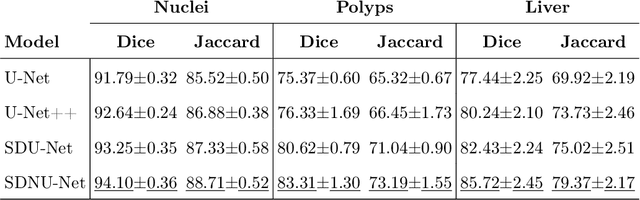
Abstract:In clinical practice, regions of interest in medical imaging often need to be identified through a process of precise image segmentation. The quality of this image segmentation step critically affects the subsequent clinical assessment of the patient status. To enable high accuracy, automatic image segmentation, we introduce a novel deep neural network architecture that exploits the inherent spatial coherence of anatomical structures and is well equipped to capture long-range spatial dependencies in the segmented pixel/voxel space. In contrast to the state-of-the-art solutions based on convolutional layers, our approach leverages on recently introduced spatial dependency layers that have an unbounded receptive field and explicitly model the inductive bias of spatial coherence. Our method performs favourably to commonly used U-Net and U-Net++ architectures as demonstrated by improved Dice and Jaccardscore in three different medical segmentation tasks: nuclei segmentation in microscopy images, polyp segmentation in colonoscopy videos, and liver segmentation in abdominal CT scans.
Spatial Dependency Networks: Neural Layers for Improved Generative Image Modeling
Mar 16, 2021



Abstract:How to improve generative modeling by better exploiting spatial regularities and coherence in images? We introduce a novel neural network for building image generators (decoders) and apply it to variational autoencoders (VAEs). In our spatial dependency networks (SDNs), feature maps at each level of a deep neural net are computed in a spatially coherent way, using a sequential gating-based mechanism that distributes contextual information across 2-D space. We show that augmenting the decoder of a hierarchical VAE by spatial dependency layers considerably improves density estimation over baseline convolutional architectures and the state-of-the-art among the models within the same class. Furthermore, we demonstrate that SDN can be applied to large images by synthesizing samples of high quality and coherence. In a vanilla VAE setting, we find that a powerful SDN decoder also improves learning disentangled representations, indicating that neural architectures play an important role in this task. Our results suggest favoring spatial dependency over convolutional layers in various VAE settings. The accompanying source code is given at https://github.com/djordjemila/sdn.
On the Transfer of Inductive Bias from Simulation to the Real World: a New Disentanglement Dataset
Jun 07, 2019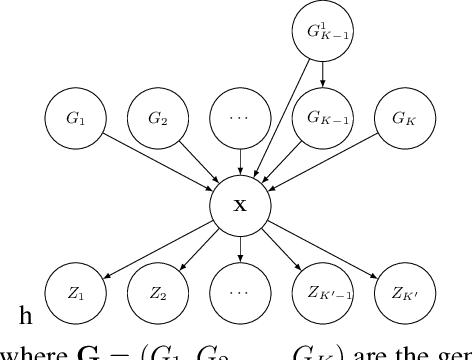
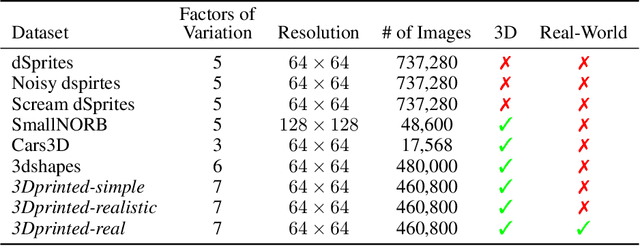
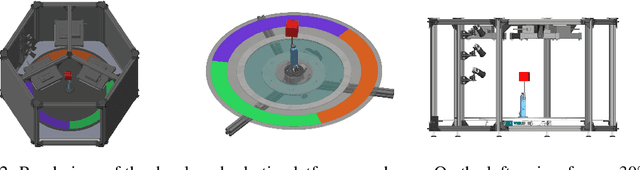
Abstract:Learning meaningful and compact representations with structurally disentangled semantic aspects is considered to be of key importance in representation learning. Since real-world data is notoriously costly to collect, many recent state-of-the-art disentanglement models have heavily relied on synthetic toy data-sets. In this paper, we propose a novel data-set which consists of over 450'000 images of physical 3D objects with seven factors of variation, such as object color, shape, size and position. In order to be able to control all the factors of variation precisely, we built an experimental platform where the objects are being moved by a robotic arm. In addition, we provide two more datasets which consist of simulations of the experimental setup. These datasets provide for the first time the possibility to systematically investigate how well different disentanglement methods perform on real data in comparison to simulation, and how simulated data can be leveraged to build better representations of the real world.
Disentangled State Space Representations
Jun 07, 2019
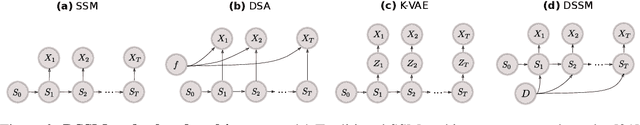
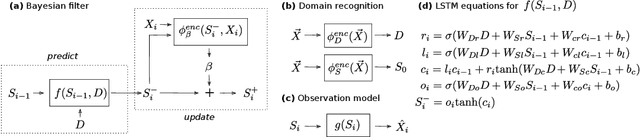
Abstract:Sequential data often originates from diverse domains across which statistical regularities and domain specifics exist. To specifically learn cross-domain sequence representations, we introduce disentangled state space models (DSSM) -- a class of SSM in which domain-invariant state dynamics is explicitly disentangled from domain-specific information governing that dynamics. We analyze how such separation can improve knowledge transfer to new domains, and enable robust prediction, sequence manipulation and domain characterization. We furthermore propose an unsupervised VAE-based training procedure to implement DSSM in form of Bayesian filters. In our experiments, we applied VAE-DSSM framework to achieve competitive performance in online ODE system identification and regression across experimental settings, and controlled generation and prediction of bouncing ball video sequences across varying gravitational influences.
Interventional Robustness of Deep Latent Variable Models
Oct 31, 2018



Abstract:The ability to learn disentangled representations that split underlying sources of variation in high dimensional, unstructured data is of central importance for data efficient and robust use of neural networks. Various approaches aiming towards this goal have been proposed in the recent time -- validating existing work is hence a crucial task to guide further development. Previous validation methods focused on shared information between generative factors and learned features. The effects of rare events or cumulative influences from multiple factors on encodings, however, remain uncaptured. Our experiments show that this already becomes noticeable in a simple, noise free dataset. This is why we introduce the interventional robustness score, which provides a quantitative evaluation of robustness in learned representations with respect to interventions on generative factors and changing nuisance factors. We show how this score can be estimated from labeled observational data, that may be confounded, and further provide an efficient algorithm that scales linearly in the dataset size. The benefits of our causally motivated framework are illustrated in extensive experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge