Liver Segmentation
Papers and Code
Towards Segmenting the Invisible: An End-to-End Registration and Segmentation Framework for Weakly Supervised Tumour Analysis
Feb 05, 2026Liver tumour ablation presents a significant clinical challenge: whilst tumours are clearly visible on pre-operative MRI, they are often effectively invisible on intra-operative CT due to minimal contrast between pathological and healthy tissue. This work investigates the feasibility of cross-modality weak supervision for scenarios where pathology is visible in one modality (MRI) but absent in another (CT). We present a hybrid registration-segmentation framework that combines MSCGUNet for inter-modal image registration with a UNet-based segmentation module, enabling registration-assisted pseudo-label generation for CT images. Our evaluation on the CHAOS dataset demonstrates that the pipeline can successfully register and segment healthy liver anatomy, achieving a Dice score of 0.72. However, when applied to clinical data containing tumours, performance degrades substantially (Dice score of 0.16), revealing the fundamental limitations of current registration methods when the target pathology lacks corresponding visual features in the target modality. We analyse the "domain gap" and "feature absence" problems, demonstrating that whilst spatial propagation of labels via registration is feasible for visible structures, segmenting truly invisible pathology remains an open challenge. Our findings highlight that registration-based label transfer cannot compensate for the absence of discriminative features in the target modality, providing important insights for future research in cross-modality medical image analysis. Code an weights are available at: https://github.com/BudhaTronix/Weakly-Supervised-Tumour-Detection
* Accepted for AIBio at ECAI 2025
Toxicity Assessment in Preclinical Histopathology via Class-Aware Mahalanobis Distance for Known and Novel Anomalies
Feb 02, 2026Drug-induced toxicity remains a leading cause of failure in preclinical development and early clinical trials. Detecting adverse effects at an early stage is critical to reduce attrition and accelerate the development of safe medicines. Histopathological evaluation remains the gold standard for toxicity assessment, but it relies heavily on expert pathologists, creating a bottleneck for large-scale screening. To address this challenge, we introduce an AI-based anomaly detection framework for histopathological whole-slide images (WSIs) in rodent livers from toxicology studies. The system identifies healthy tissue and known pathologies (anomalies) for which training data is available. In addition, it can detect rare pathologies without training data as out-of-distribution (OOD) findings. We generate a novel dataset of pixelwise annotations of healthy tissue and known pathologies and use this data to fine-tune a pre-trained Vision Transformer (DINOv2) via Low-Rank Adaptation (LoRA) in order to do tissue segmentation. Finally, we extract features for OOD detection using the Mahalanobis distance. To better account for class-dependent variability in histological data, we propose the use of class-specific thresholds. We optimize the thresholds using the mean of the false negative and false positive rates, resulting in only 0.16\% of pathological tissue classified as healthy and 0.35\% of healthy tissue classified as pathological. Applied to mouse liver WSIs with known toxicological findings, the framework accurately detects anomalies, including rare OOD morphologies. This work demonstrates the potential of AI-driven histopathology to support preclinical workflows, reduce late-stage failures, and improve efficiency in drug development.
A-QCF-Net: An Adaptive Quaternion Cross-Fusion Network for Multimodal Liver Tumor Segmentation from Unpaired Datasets
Dec 25, 2025



Multimodal medical imaging provides complementary information that is crucial for accurate delineation of pathology, but the development of deep learning models is limited by the scarcity of large datasets in which different modalities are paired and spatially aligned. This paper addresses this fundamental limitation by proposing an Adaptive Quaternion Cross-Fusion Network (A-QCF-Net) that learns a single unified segmentation model from completely separate and unpaired CT and MRI cohorts. The architecture exploits the parameter efficiency and expressive power of Quaternion Neural Networks to construct a shared feature space. At its core is the Adaptive Quaternion Cross-Fusion (A-QCF) block, a data driven attention module that enables bidirectional knowledge transfer between the two streams. By learning to modulate the flow of information dynamically, the A-QCF block allows the network to exchange abstract modality specific expertise, such as the sharp anatomical boundary information available in CT and the subtle soft tissue contrast provided by MRI. This mutual exchange regularizes and enriches the feature representations of both streams. We validate the framework by jointly training a single model on the unpaired LiTS (CT) and ATLAS (MRI) datasets. The jointly trained model achieves Tumor Dice scores of 76.7% on CT and 78.3% on MRI, significantly exceeding the strong unimodal nnU-Net baseline by margins of 5.4% and 4.7% respectively. Furthermore, comprehensive explainability analysis using Grad-CAM and Grad-CAM++ confirms that the model correctly focuses on relevant pathological structures, ensuring the learned representations are clinically meaningful. This provides a robust and clinically viable paradigm for unlocking the large unpaired imaging archives that are common in healthcare.
Liver Fibrosis Quantification and Analysis: The LiQA Dataset and Baseline Method
Dec 22, 2025Liver fibrosis represents a significant global health burden, necessitating accurate staging for effective clinical management. This report introduces the LiQA (Liver Fibrosis Quantification and Analysis) dataset, established as part of the CARE 2024 challenge. Comprising $440$ patients with multi-phase, multi-center MRI scans, the dataset is curated to benchmark algorithms for Liver Segmentation (LiSeg) and Liver Fibrosis Staging (LiFS) under complex real-world conditions, including domain shifts, missing modalities, and spatial misalignment. We further describe the challenge's top-performing methodology, which integrates a semi-supervised learning framework with external data for robust segmentation, and utilizes a multi-view consensus approach with Class Activation Map (CAM)-based regularization for staging. Evaluation of this baseline demonstrates that leveraging multi-source data and anatomical constraints significantly enhances model robustness in clinical settings.
SSL-MedSAM2: A Semi-supervised Medical Image Segmentation Framework Powered by Few-shot Learning of SAM2
Dec 12, 2025Despite the success of deep learning based models in medical image segmentation, most state-of-the-art (SOTA) methods perform fully-supervised learning, which commonly rely on large scale annotated training datasets. However, medical image annotation is highly time-consuming, hindering its clinical applications. Semi-supervised learning (SSL) has been emerged as an appealing strategy in training with limited annotations, largely reducing the labelling cost. We propose a novel SSL framework SSL-MedSAM2, which contains a training-free few-shot learning branch TFFS-MedSAM2 based on the pretrained large foundation model Segment Anything Model 2 (SAM2) for pseudo label generation, and an iterative fully-supervised learning branch FSL-nnUNet based on nnUNet for pseudo label refinement. The results on MICCAI2025 challenge CARE-LiSeg (Liver Segmentation) demonstrate an outstanding performance of SSL-MedSAM2 among other methods. The average dice scores on the test set in GED4 and T1 MRI are 0.9710 and 0.9648 respectively, and the Hausdorff distances are 20.07 and 21.97 respectively. The code is available via https://github.com/naisops/SSL-MedSAM2/tree/main.
Precise Liver Tumor Segmentation in CT Using a Hybrid Deep Learning-Radiomics Framework
Dec 08, 2025Accurate three-dimensional delineation of liver tumors on contrast-enhanced CT is a prerequisite for treatment planning, navigation and response assessment, yet manual contouring is slow, observer-dependent and difficult to standardise across centres. Automatic segmentation is complicated by low lesion-parenchyma contrast, blurred or incomplete boundaries, heterogeneous enhancement patterns, and confounding structures such as vessels and adjacent organs. We propose a hybrid framework that couples an attention-enhanced cascaded U-Net with handcrafted radiomics and voxel-wise 3D CNN refinement for joint liver and liver-tumor segmentation. First, a 2.5D two-stage network with a densely connected encoder, sub-pixel convolution decoders and multi-scale attention gates produces initial liver and tumor probability maps from short stacks of axial slices. Inter-slice temporal consistency is then enforced by a simple three-slice refinement rule along the cranio-caudal direction, which restores thin and tiny lesions while suppressing isolated noise. Next, 728 radiomic descriptors spanning intensity, texture, shape, boundary and wavelet feature groups are extracted from candidate lesions and reduced to 20 stable, highly informative features via multi-strategy feature selection; a random forest classifier uses these features to reject false-positive regions. Finally, a compact 3D patch-based CNN derived from AlexNet operates in a narrow band around the tumor boundary to perform voxel-level relabelling and contour smoothing.
FocusSDF: Boundary-Aware Learning for Medical Image Segmentation via Signed Distance Supervision
Nov 14, 2025Segmentation of medical images constitutes an essential component of medical image analysis, providing the foundation for precise diagnosis and efficient therapeutic interventions in clinical practices. Despite substantial progress, most segmentation models do not explicitly encode boundary information; as a result, making boundary preservation a persistent challenge in medical image segmentation. To address this challenge, we introduce FocusSDF, a novel loss function based on the signed distance functions (SDFs), which redirects the network to concentrate on boundary regions by adaptively assigning higher weights to pixels closer to the lesion or organ boundary, effectively making it boundary aware. To rigorously validate FocusSDF, we perform extensive evaluations against five state-of-the-art medical image segmentation models, including the foundation model MedSAM, using four distance-based loss functions across diverse datasets covering cerebral aneurysm, stroke, liver, and breast tumor segmentation tasks spanning multiple imaging modalities. The experimental results consistently demonstrate the superior performance of FocusSDF over existing distance transform based loss functions.
Task-Adaptive Low-Dose CT Reconstruction
Nov 10, 2025



Deep learning-based low-dose computed tomography reconstruction methods already achieve high performance on standard image quality metrics like peak signal-to-noise ratio and structural similarity index measure. Yet, they frequently fail to preserve the critical anatomical details needed for diagnostic tasks. This fundamental limitation hinders their clinical applicability despite their high metric scores. We propose a novel task-adaptive reconstruction framework that addresses this gap by incorporating a frozen pre-trained task network as a regularization term in the reconstruction loss function. Unlike existing joint-training approaches that simultaneously optimize both reconstruction and task networks, and risk diverging from satisfactory reconstructions, our method leverages a pre-trained task model to guide reconstruction training while still maintaining diagnostic quality. We validate our framework on a liver and liver tumor segmentation task. Our task-adaptive models achieve Dice scores up to 0.707, approaching the performance of full-dose scans (0.874), and substantially outperforming joint-training approaches (0.331) and traditional reconstruction methods (0.626). Critically, our framework can be integrated into any existing deep learning-based reconstruction model through simple loss function modification, enabling widespread adoption for task-adaptive optimization in clinical practice. Our codes are available at: https://github.com/itu-biai/task_adaptive_ct
Automatic segmentation of colorectal liver metastases for ultrasound-based navigated resection
Nov 07, 2025


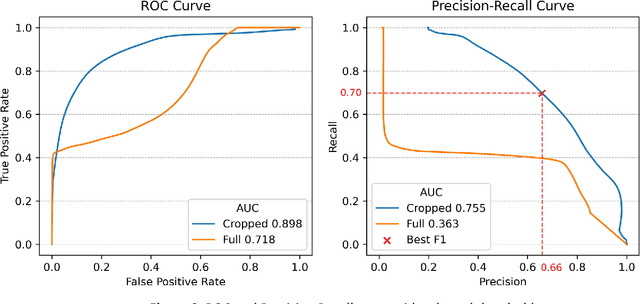
Introduction: Accurate intraoperative delineation of colorectal liver metastases (CRLM) is crucial for achieving negative resection margins but remains challenging using intraoperative ultrasound (iUS) due to low contrast, noise, and operator dependency. Automated segmentation could enhance precision and efficiency in ultrasound-based navigation workflows. Methods: Eighty-five tracked 3D iUS volumes from 85 CRLM patients were used to train and evaluate a 3D U-Net implemented via the nnU-Net framework. Two variants were compared: one trained on full iUS volumes and another on cropped regions around tumors. Segmentation accuracy was assessed using Dice Similarity Coefficient (DSC), Hausdorff Distance (HDist.), and Relative Volume Difference (RVD) on retrospective and prospective datasets. The workflow was integrated into 3D Slicer for real-time intraoperative use. Results: The cropped-volume model significantly outperformed the full-volume model across all metrics (AUC-ROC = 0.898 vs 0.718). It achieved median DSC = 0.74, recall = 0.79, and HDist. = 17.1 mm comparable to semi-automatic segmentation but with ~4x faster execution (~ 1 min). Prospective intraoperative testing confirmed robust and consistent performance, with clinically acceptable accuracy for real-time surgical guidance. Conclusion: Automatic 3D segmentation of CRLM in iUS using a cropped 3D U-Net provides reliable, near real-time results with minimal operator input. The method enables efficient, registration-free ultrasound-based navigation for hepatic surgery, approaching expert-level accuracy while substantially reducing manual workload and procedure time.
Label-Efficient Cross-Modality Generalization for Liver Segmentation in Multi-Phase MRI
Oct 06, 2025Accurate liver segmentation in multi-phase MRI is vital for liver fibrosis assessment, yet labeled data is often scarce and unevenly distributed across imaging modalities and vendor systems. We propose a label-efficient segmentation approach that promotes cross-modality generalization under real-world conditions, where GED4 hepatobiliary-phase annotations are limited, non-contrast sequences (T1WI, T2WI, DWI) are unlabeled, and spatial misalignment and missing phases are common. Our method integrates a foundation-scale 3D segmentation backbone adapted via fine-tuning, co-training with cross pseudo supervision to leverage unlabeled volumes, and a standardized preprocessing pipeline. Without requiring spatial registration, the model learns to generalize across MRI phases and vendors, demonstrating robust segmentation performance in both labeled and unlabeled domains. Our results exhibit the effectiveness of our proposed label-efficient baseline for liver segmentation in multi-phase, multi-vendor MRI and highlight the potential of combining foundation model adaptation with co-training for real-world clinical imaging tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge