Chirag Mandal
Towards Segmenting the Invisible: An End-to-End Registration and Segmentation Framework for Weakly Supervised Tumour Analysis
Feb 05, 2026Abstract:Liver tumour ablation presents a significant clinical challenge: whilst tumours are clearly visible on pre-operative MRI, they are often effectively invisible on intra-operative CT due to minimal contrast between pathological and healthy tissue. This work investigates the feasibility of cross-modality weak supervision for scenarios where pathology is visible in one modality (MRI) but absent in another (CT). We present a hybrid registration-segmentation framework that combines MSCGUNet for inter-modal image registration with a UNet-based segmentation module, enabling registration-assisted pseudo-label generation for CT images. Our evaluation on the CHAOS dataset demonstrates that the pipeline can successfully register and segment healthy liver anatomy, achieving a Dice score of 0.72. However, when applied to clinical data containing tumours, performance degrades substantially (Dice score of 0.16), revealing the fundamental limitations of current registration methods when the target pathology lacks corresponding visual features in the target modality. We analyse the "domain gap" and "feature absence" problems, demonstrating that whilst spatial propagation of labels via registration is feasible for visible structures, segmenting truly invisible pathology remains an open challenge. Our findings highlight that registration-based label transfer cannot compensate for the absence of discriminative features in the target modality, providing important insights for future research in cross-modality medical image analysis. Code an weights are available at: https://github.com/BudhaTronix/Weakly-Supervised-Tumour-Detection
* Accepted for AIBio at ECAI 2025
TorchEsegeta: Framework for Interpretability and Explainability of Image-based Deep Learning Models
Oct 16, 2021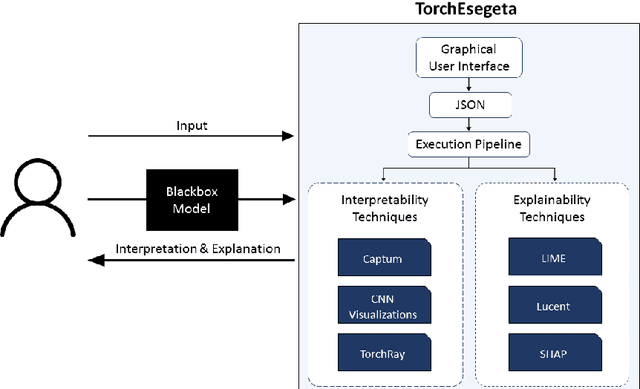
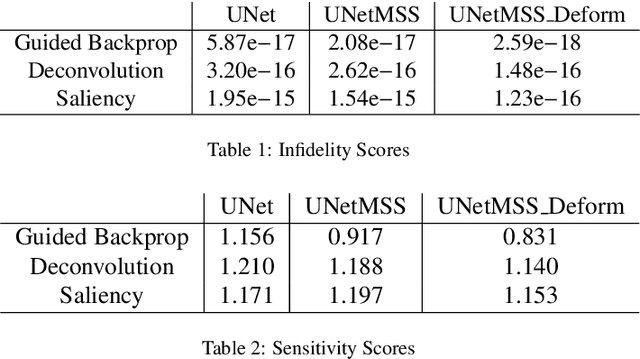
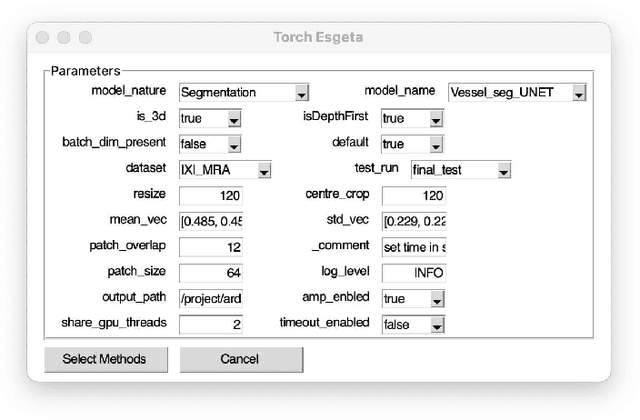
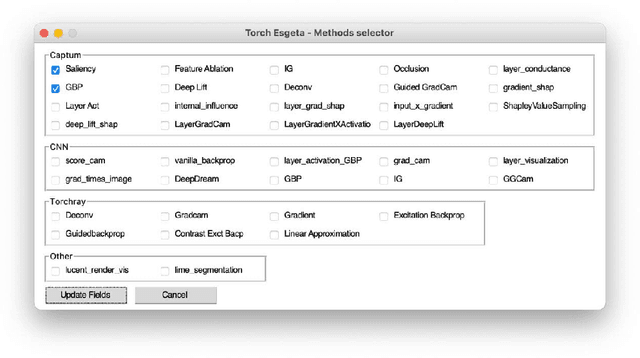
Abstract:Clinicians are often very sceptical about applying automatic image processing approaches, especially deep learning based methods, in practice. One main reason for this is the black-box nature of these approaches and the inherent problem of missing insights of the automatically derived decisions. In order to increase trust in these methods, this paper presents approaches that help to interpret and explain the results of deep learning algorithms by depicting the anatomical areas which influence the decision of the algorithm most. Moreover, this research presents a unified framework, TorchEsegeta, for applying various interpretability and explainability techniques for deep learning models and generate visual interpretations and explanations for clinicians to corroborate their clinical findings. In addition, this will aid in gaining confidence in such methods. The framework builds on existing interpretability and explainability techniques that are currently focusing on classification models, extending them to segmentation tasks. In addition, these methods have been adapted to 3D models for volumetric analysis. The proposed framework provides methods to quantitatively compare visual explanations using infidelity and sensitivity metrics. This framework can be used by data scientists to perform post-hoc interpretations and explanations of their models, develop more explainable tools and present the findings to clinicians to increase their faith in such models. The proposed framework was evaluated based on a use case scenario of vessel segmentation models trained on Time-of-fight (TOF) Magnetic Resonance Angiogram (MRA) images of the human brain. Quantitative and qualitative results of a comparative study of different models and interpretability methods are presented. Furthermore, this paper provides an extensive overview of several existing interpretability and explainability methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge