Zhongxi Qiu
Open-Medical-R1: How to Choose Data for RLVR Training at Medicine Domain
Apr 16, 2025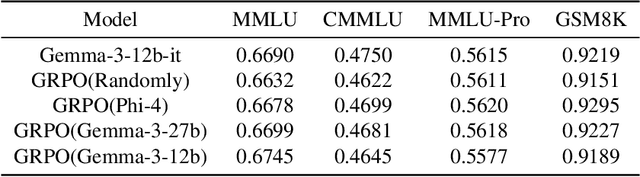
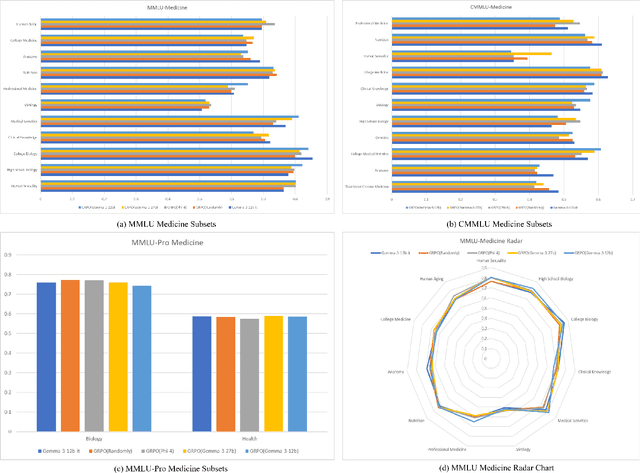
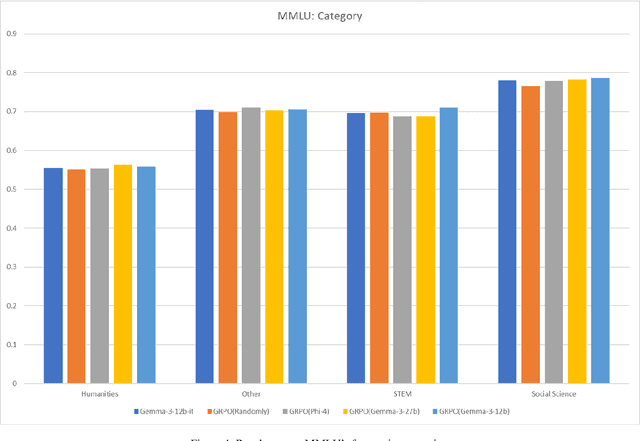
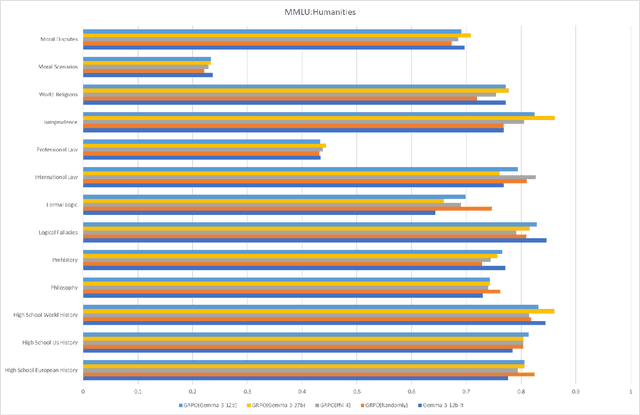
Abstract:This paper explores optimal data selection strategies for Reinforcement Learning with Verified Rewards (RLVR) training in the medical domain. While RLVR has shown exceptional potential for enhancing reasoning capabilities in large language models, most prior implementations have focused on mathematics and logical puzzles, with limited exploration of domain-specific applications like medicine. We investigate four distinct data sampling strategies from MedQA-USMLE: random sampling (baseline), and filtering using Phi-4, Gemma-3-27b-it, and Gemma-3-12b-it models. Using Gemma-3-12b-it as our base model and implementing Group Relative Policy Optimization (GRPO), we evaluate performance across multiple benchmarks including MMLU, GSM8K, MMLU-Pro, and CMMLU. Our findings demonstrate that models trained on filtered data generally outperform those trained on randomly selected samples. Notably, training on self-filtered samples (using Gemma-3-12b-it for filtering) achieved superior performance in medical domains but showed reduced robustness across different benchmarks, while filtering with larger models from the same series yielded better overall robustness. These results provide valuable insights into effective data organization strategies for RLVR in specialized domains and highlight the importance of thoughtful data selection in achieving optimal performance. You can access our repository (https://github.com/Qsingle/open-medical-r1) to get the codes.
COph100: A comprehensive fundus image registration dataset from infants constituting the "RIDIRP" database
Jan 06, 2025Abstract:Retinal image registration is vital for diagnostic therapeutic applications within the field of ophthalmology. Existing public datasets, focusing on adult retinal pathologies with high-quality images, have limited number of image pairs and neglect clinical challenges. To address this gap, we introduce COph100, a novel and challenging dataset known as the Comprehensive Ophthalmology Retinal Image Registration dataset for infants with a wide range of image quality issues constituting the public "RIDIRP" database. COph100 consists of 100 eyes, each with 2 to 9 examination sessions, amounting to a total of 491 image pairs carefully selected from the publicly available dataset. We manually labeled the corresponding ground truth image points and provided automatic vessel segmentation masks for each image. We have assessed COph100 in terms of image quality and registration outcomes using state-of-the-art algorithms. This resource enables a robust comparison of retinal registration methodologies and aids in the analysis of disease progression in infants, thereby deepening our understanding of pediatric ophthalmic conditions.
* 12 pages, 7 figures
RaffeSDG: Random Frequency Filtering enabled Single-source Domain Generalization for Medical Image Segmentation
May 02, 2024



Abstract:Deep learning models often encounter challenges in making accurate inferences when there are domain shifts between the source and target data. This issue is particularly pronounced in clinical settings due to the scarcity of annotated data resulting from the professional and private nature of medical data. Despite the existence of decent solutions, many of them are hindered in clinical settings due to limitations in data collection and computational complexity. To tackle domain shifts in data-scarce medical scenarios, we propose a Random frequency filtering enabled Single-source Domain Generalization algorithm (RaffeSDG), which promises robust out-of-domain inference with segmentation models trained on a single-source domain. A filter-based data augmentation strategy is first proposed to promote domain variability within a single-source domain by introducing variations in frequency space and blending homologous samples. Then Gaussian filter-based structural saliency is also leveraged to learn robust representations across augmented samples, further facilitating the training of generalizable segmentation models. To validate the effectiveness of RaffeSDG, we conducted extensive experiments involving out-of-domain inference on segmentation tasks for three human tissues imaged by four diverse modalities. Through thorough investigations and comparisons, compelling evidence was observed in these experiments, demonstrating the potential and generalizability of RaffeSDG. The code is available at https://github.com/liamheng/Non-IID_Medical_Image_Segmentation.
Enhancing and Adapting in the Clinic: Source-free Unsupervised Domain Adaptation for Medical Image Enhancement
Dec 03, 2023Abstract:Medical imaging provides many valuable clues involving anatomical structure and pathological characteristics. However, image degradation is a common issue in clinical practice, which can adversely impact the observation and diagnosis by physicians and algorithms. Although extensive enhancement models have been developed, these models require a well pre-training before deployment, while failing to take advantage of the potential value of inference data after deployment. In this paper, we raise an algorithm for source-free unsupervised domain adaptive medical image enhancement (SAME), which adapts and optimizes enhancement models using test data in the inference phase. A structure-preserving enhancement network is first constructed to learn a robust source model from synthesized training data. Then a teacher-student model is initialized with the source model and conducts source-free unsupervised domain adaptation (SFUDA) by knowledge distillation with the test data. Additionally, a pseudo-label picker is developed to boost the knowledge distillation of enhancement tasks. Experiments were implemented on ten datasets from three medical image modalities to validate the advantage of the proposed algorithm, and setting analysis and ablation studies were also carried out to interpret the effectiveness of SAME. The remarkable enhancement performance and benefits for downstream tasks demonstrate the potential and generalizability of SAME. The code is available at https://github.com/liamheng/Annotation-free-Medical-Image-Enhancement.
Learnable Ophthalmology SAM
Apr 26, 2023Abstract:Segmentation is vital for ophthalmology image analysis. But its various modal images hinder most of the existing segmentation algorithms applications, as they rely on training based on a large number of labels or hold weak generalization ability. Based on Segment Anything (SAM), we propose a simple but effective learnable prompt layer suitable for multiple target segmentation in ophthalmology multi-modal images, named Learnable Ophthalmology Segment Anything (SAM). The learnable prompt layer learns medical prior knowledge from each transformer layer. During training, we only train the prompt layer and task head based on a one-shot mechanism. We demonstrate the effectiveness of our thought based on four medical segmentation tasks based on nine publicly available datasets. Moreover, we only provide a new improvement thought for applying the existing fundamental CV models in the medical field. Our codes are available at \href{https://github.com/Qsingle/LearnablePromptSAM}{website}.
Hard Exudate Segmentation Supplemented by Super-Resolution with Multi-scale Attention Fusion Module
Nov 17, 2022Abstract:Hard exudates (HE) is the most specific biomarker for retina edema. Precise HE segmentation is vital for disease diagnosis and treatment, but automatic segmentation is challenged by its large variation of characteristics including size, shape and position, which makes it difficult to detect tiny lesions and lesion boundaries. Considering the complementary features between segmentation and super-resolution tasks, this paper proposes a novel hard exudates segmentation method named SS-MAF with an auxiliary super-resolution task, which brings in helpful detailed features for tiny lesion and boundaries detection. Specifically, we propose a fusion module named Multi-scale Attention Fusion (MAF) module for our dual-stream framework to effectively integrate features of the two tasks. MAF first adopts split spatial convolutional (SSC) layer for multi-scale features extraction and then utilize attention mechanism for features fusion of the two tasks. Considering pixel dependency, we introduce region mutual information (RMI) loss to optimize MAF module for tiny lesions and boundary detection. We evaluate our method on two public lesion datasets, IDRiD and E-Ophtha. Our method shows competitive performance with low-resolution inputs, both quantitatively and qualitatively. On E-Ophtha dataset, the method can achieve $\geq3\%$ higher dice and recall compared with the state-of-the-art methods.
SuperVessel: Segmenting High-resolution Vessel from Low-resolution Retinal Image
Jul 28, 2022



Abstract:Vascular segmentation extracts blood vessels from images and serves as the basis for diagnosing various diseases, like ophthalmic diseases. Ophthalmologists often require high-resolution segmentation results for analysis, which leads to super-computational load by most existing methods. If based on low-resolution input, they easily ignore tiny vessels or cause discontinuity of segmented vessels. To solve these problems, the paper proposes an algorithm named SuperVessel, which gives out high-resolution and accurate vessel segmentation using low-resolution images as input. We first take super-resolution as our auxiliary branch to provide potential high-resolution detail features, which can be deleted in the test phase. Secondly, we propose two modules to enhance the features of the interested segmentation region, including an upsampling with feature decomposition (UFD) module and a feature interaction module (FIM) with a constraining loss to focus on the interested features. Extensive experiments on three publicly available datasets demonstrate that our proposed SuperVessel can segment more tiny vessels with higher segmentation accuracy IoU over 6%, compared with other state-of-the-art algorithms. Besides, the stability of SuperVessel is also stronger than other algorithms. We will release the code after the paper is published.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge