Zheng Feng
Automatic News Generation and Fact-Checking System Based on Language Processing
May 17, 2024Abstract:This paper explores an automatic news generation and fact-checking system based on language processing, aimed at enhancing the efficiency and quality of news production while ensuring the authenticity and reliability of the news content. With the rapid development of Natural Language Processing (NLP) and deep learning technologies, automatic news generation systems are capable of extracting key information from massive data and generating well-structured, fluent news articles. Meanwhile, by integrating fact-checking technology, the system can effectively prevent the spread of false news and improve the accuracy and credibility of news. This study details the key technologies involved in automatic news generation and factchecking, including text generation, information extraction, and the application of knowledge graphs, and validates the effectiveness of these technologies through experiments. Additionally, the paper discusses the future development directions of automatic news generation and fact-checking systems, emphasizing the importance of further integration and innovation of technologies. The results show that with continuous technological optimization and practical application, these systems will play an increasingly important role in the future news industry, providing more efficient and reliable news services.
DR-VIDAL -- Doubly Robust Variational Information-theoretic Deep Adversarial Learning for Counterfactual Prediction and Treatment Effect Estimation on Real World Data
Mar 07, 2023Abstract:Determining causal effects of interventions onto outcomes from real-world, observational (non-randomized) data, e.g., treatment repurposing using electronic health records, is challenging due to underlying bias. Causal deep learning has improved over traditional techniques for estimating individualized treatment effects (ITE). We present the Doubly Robust Variational Information-theoretic Deep Adversarial Learning (DR-VIDAL), a novel generative framework that combines two joint models of treatment and outcome, ensuring an unbiased ITE estimation even when one of the two is misspecified. DR-VIDAL integrates: (i) a variational autoencoder (VAE) to factorize confounders into latent variables according to causal assumptions; (ii) an information-theoretic generative adversarial network (Info-GAN) to generate counterfactuals; (iii) a doubly robust block incorporating treatment propensities for outcome predictions. On synthetic and real-world datasets (Infant Health and Development Program, Twin Birth Registry, and National Supported Work Program), DR-VIDAL achieves better performance than other non-generative and generative methods. In conclusion, DR-VIDAL uniquely fuses causal assumptions, VAE, Info-GAN, and doubly robustness into a comprehensive, performant framework. Code is available at: https://github.com/Shantanu48114860/DR-VIDAL-AMIA-22 under MIT license.
BatmanNet: Bi-branch Masked Graph Transformer Autoencoder for Molecular Representation
Nov 29, 2022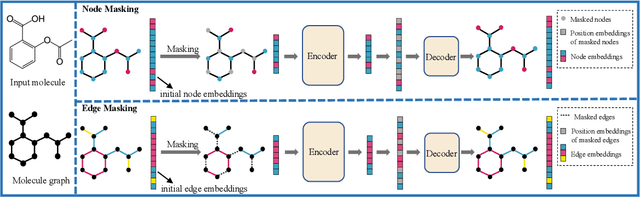
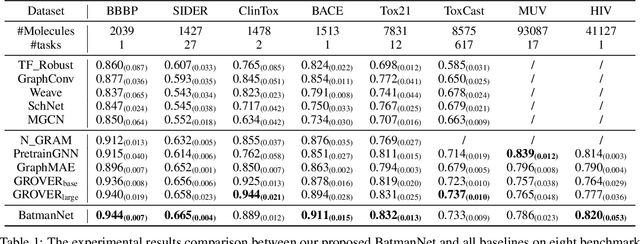
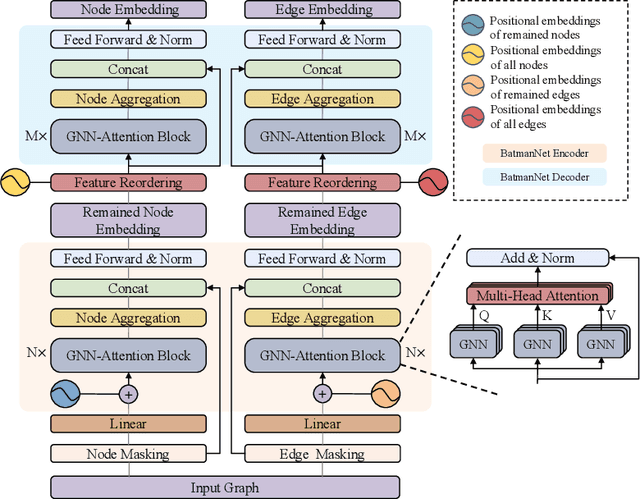
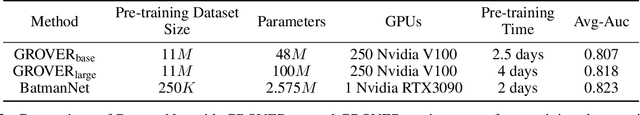
Abstract:Although substantial efforts have been made using graph neural networks (GNNs) for AI-driven drug discovery (AIDD), effective molecular representation learning remains an open challenge, especially in the case of insufficient labeled molecules. Recent studies suggest that big GNN models pre-trained by self-supervised learning on unlabeled datasets enable better transfer performance in downstream molecular property prediction tasks. However, they often require large-scale datasets and considerable computational resources, which is time-consuming, computationally expensive, and environmentally unfriendly. To alleviate these limitations, we propose a novel pre-training model for molecular representation learning, Bi-branch Masked Graph Transformer Autoencoder (BatmanNet). BatmanNet features two tailored and complementary graph autoencoders to reconstruct the missing nodes and edges from a masked molecular graph. To our surprise, BatmanNet discovered that the highly masked proportion (60%) of the atoms and bonds achieved the best performance. We further propose an asymmetric graph-based encoder-decoder architecture for either nodes and edges, where a transformer-based encoder only takes the visible subset of nodes or edges, and a lightweight decoder reconstructs the original molecule from the latent representation and mask tokens. With this simple yet effective asymmetrical design, our BatmanNet can learn efficiently even from a much smaller-scale unlabeled molecular dataset to capture the underlying structural and semantic information, overcoming a major limitation of current deep neural networks for molecular representation learning. For instance, using only 250K unlabelled molecules as pre-training data, our BatmanNet with 2.575M parameters achieves a 0.5% improvement on the average AUC compared with the current state-of-the-art method with 100M parameters pre-trained on 11M molecules.
Variational Temporal Deconfounder for Individualized Treatment Effect Estimation from Longitudinal Observational Data
Jul 23, 2022


Abstract:Estimating treatment effects, especially individualized treatment effects (ITE), using observational data is challenging due to the complex situations of confounding bias. Existing approaches for estimating treatment effects from longitudinal observational data are usually built upon a strong assumption of "unconfoundedness", which is hard to fulfill in real-world practice. In this paper, we propose the Variational Temporal Deconfounder (VTD), an approach that leverages deep variational embeddings in the longitudinal setting using proxies (i.e., surrogate variables that serve for unobservable variables). Specifically, VTD leverages observed proxies to learn a hidden embedding that reflects the true hidden confounders in the observational data. As such, our VTD method does not rely on the "unconfoundedness" assumption. We test our VTD method on both synthetic and real-world clinical data, and the results show that our approach is effective when hidden confounding is the leading bias compared to other existing models.
Generalized Batch Normalization: Towards Accelerating Deep Neural Networks
Dec 08, 2018



Abstract:Utilizing recently introduced concepts from statistics and quantitative risk management, we present a general variant of Batch Normalization (BN) that offers accelerated convergence of Neural Network training compared to conventional BN. In general, we show that mean and standard deviation are not always the most appropriate choice for the centering and scaling procedure within the BN transformation, particularly if ReLU follows the normalization step. We present a Generalized Batch Normalization (GBN) transformation, which can utilize a variety of alternative deviation measures for scaling and statistics for centering, choices which naturally arise from the theory of generalized deviation measures and risk theory in general. When used in conjunction with the ReLU non-linearity, the underlying risk theory suggests natural, arguably optimal choices for the deviation measure and statistic. Utilizing the suggested deviation measure and statistic, we show experimentally that training is accelerated more so than with conventional BN, often with improved error rate as well. Overall, we propose a more flexible BN transformation supported by a complimentary theoretical framework that can potentially guide design choices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge