Shantanu Ghosh
Semantic Consistency-Based Uncertainty Quantification for Factuality in Radiology Report Generation
Dec 05, 2024



Abstract:Radiology report generation (RRG) has shown great potential in assisting radiologists by automating the labor-intensive task of report writing. While recent advancements have improved the quality and coherence of generated reports, ensuring their factual correctness remains a critical challenge. Although generative medical Vision Large Language Models (VLLMs) have been proposed to address this issue, these models are prone to hallucinations and can produce inaccurate diagnostic information. To address these concerns, we introduce a novel Semantic Consistency-Based Uncertainty Quantification framework that provides both report-level and sentence-level uncertainties. Unlike existing approaches, our method does not require modifications to the underlying model or access to its inner state, such as output token logits, thus serving as a plug-and-play module that can be seamlessly integrated with state-of-the-art models. Extensive experiments demonstrate the efficacy of our method in detecting hallucinations and enhancing the factual accuracy of automatically generated radiology reports. By abstaining from high-uncertainty reports, our approach improves factuality scores by $10$%, achieved by rejecting $20$% of reports using the Radialog model on the MIMIC-CXR dataset. Furthermore, sentence-level uncertainty flags the lowest-precision sentence in each report with an $82.9$% success rate.
Distributionally robust self-supervised learning for tabular data
Oct 11, 2024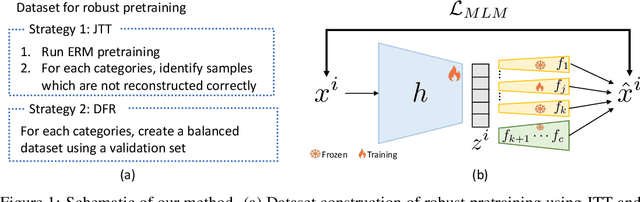
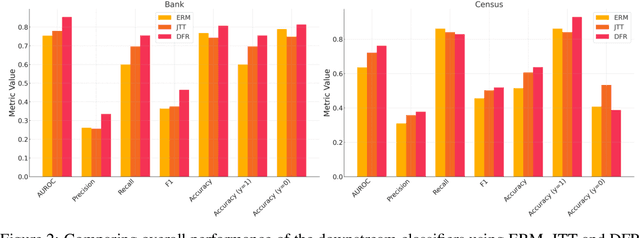
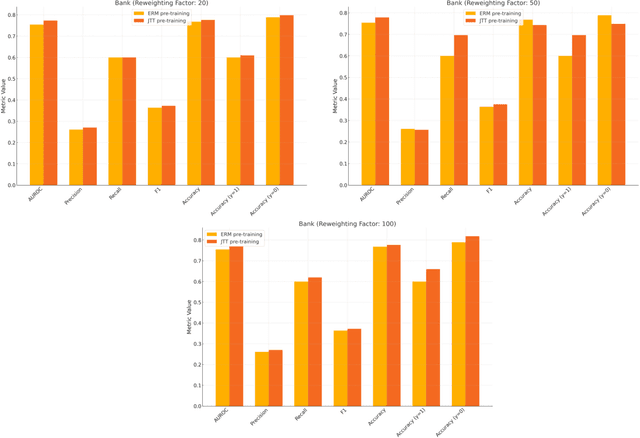
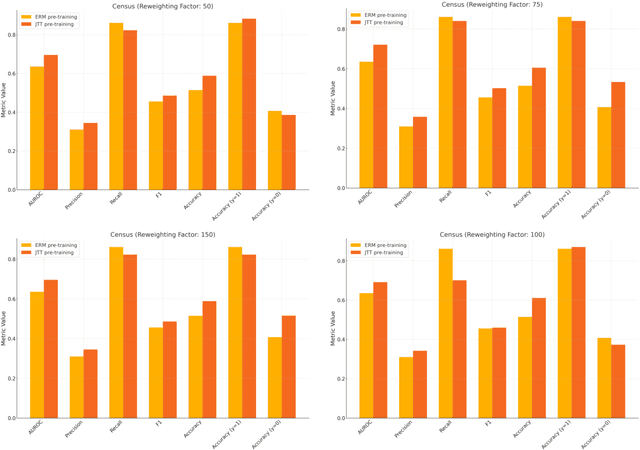
Abstract:Machine learning (ML) models trained using Empirical Risk Minimization (ERM) often exhibit systematic errors on specific subpopulations of tabular data, known as error slices. Learning robust representation in presence of error slices is challenging, especially in self-supervised settings during the feature reconstruction phase, due to high cardinality features and the complexity of constructing error sets. Traditional robust representation learning methods are largely focused on improving worst group performance in supervised setting in computer vision, leaving a gap in approaches tailored for tabular data. We address this gap by developing a framework to learn robust representation in tabular data during self-supervised pre-training. Our approach utilizes an encoder-decoder model trained with Masked Language Modeling (MLM) loss to learn robust latent representations. This paper applies the Just Train Twice (JTT) and Deep Feature Reweighting (DFR) methods during the pre-training phase for tabular data. These methods fine-tune the ERM pre-trained model by up-weighting error-prone samples or creating balanced datasets for specific categorical features. This results in specialized models for each feature, which are then used in an ensemble approach to enhance downstream classification performance. This methodology improves robustness across slices, thus enhancing overall generalization performance. Extensive experiments across various datasets demonstrate the efficacy of our approach.
Mammo-CLIP: A Vision Language Foundation Model to Enhance Data Efficiency and Robustness in Mammography
May 22, 2024Abstract:The lack of large and diverse training data on Computer-Aided Diagnosis (CAD) in breast cancer detection has been one of the concerns that impedes the adoption of the system. Recently, pre-training with large-scale image text datasets via Vision-Language models (VLM) (\eg CLIP) partially addresses the issue of robustness and data efficiency in computer vision (CV). This paper proposes Mammo-CLIP, the first VLM pre-trained on a substantial amount of screening mammogram-report pairs, addressing the challenges of dataset diversity and size. Our experiments on two public datasets demonstrate strong performance in classifying and localizing various mammographic attributes crucial for breast cancer detection, showcasing data efficiency and robustness similar to CLIP in CV. We also propose Mammo-FActOR, a novel feature attribution method, to provide spatial interpretation of representation with sentence-level granularity within mammography reports. Code is available publicly: \url{https://github.com/batmanlab/Mammo-CLIP}.
Dividing and Conquering a BlackBox to a Mixture of Interpretable Models: Route, Interpret, Repeat
Jul 12, 2023Abstract:ML model design either starts with an interpretable model or a Blackbox and explains it post hoc. Blackbox models are flexible but difficult to explain, while interpretable models are inherently explainable. Yet, interpretable models require extensive ML knowledge and tend to be less flexible and underperforming than their Blackbox variants. This paper aims to blur the distinction between a post hoc explanation of a Blackbox and constructing interpretable models. Beginning with a Blackbox, we iteratively carve out a mixture of interpretable experts (MoIE) and a residual network. Each interpretable model specializes in a subset of samples and explains them using First Order Logic (FOL), providing basic reasoning on concepts from the Blackbox. We route the remaining samples through a flexible residual. We repeat the method on the residual network until all the interpretable models explain the desired proportion of data. Our extensive experiments show that our route, interpret, and repeat approach (1) identifies a diverse set of instance-specific concepts with high concept completeness via MoIE without compromising in performance, (2) identifies the relatively ``harder'' samples to explain via residuals, (3) outperforms the interpretable by-design models by significant margins during test-time interventions, and (4) fixes the shortcut learned by the original Blackbox. The code for MoIE is publicly available at: \url{https://github.com/batmanlab/ICML-2023-Route-interpret-repeat}
* appeared as v5 of arXiv:2302.10289 which was replaced in error, which drifted into a different work, accepted in ICML 2023
Exploring the Lottery Ticket Hypothesis with Explainability Methods: Insights into Sparse Network Performance
Jul 07, 2023Abstract:Discovering a high-performing sparse network within a massive neural network is advantageous for deploying them on devices with limited storage, such as mobile phones. Additionally, model explainability is essential to fostering trust in AI. The Lottery Ticket Hypothesis (LTH) finds a network within a deep network with comparable or superior performance to the original model. However, limited study has been conducted on the success or failure of LTH in terms of explainability. In this work, we examine why the performance of the pruned networks gradually increases or decreases. Using Grad-CAM and Post-hoc concept bottleneck models (PCBMs), respectively, we investigate the explainability of pruned networks in terms of pixels and high-level concepts. We perform extensive experiments across vision and medical imaging datasets. As more weights are pruned, the performance of the network degrades. The discovered concepts and pixels from the pruned networks are inconsistent with the original network -- a possible reason for the drop in performance.
Distilling BlackBox to Interpretable models for Efficient Transfer Learning
Jun 10, 2023Abstract:Building generalizable AI models is one of the primary challenges in the healthcare domain. While radiologists rely on generalizable descriptive rules of abnormality, Neural Network (NN) models suffer even with a slight shift in input distribution (e.g., scanner type). Fine-tuning a model to transfer knowledge from one domain to another requires a significant amount of labeled data in the target domain. In this paper, we develop an interpretable model that can be efficiently fine-tuned to an unseen target domain with minimal computational cost. We assume the interpretable component of NN to be approximately domain-invariant. However, interpretable models typically underperform compared to their Blackbox (BB) variants. We start with a BB in the source domain and distill it into a \emph{mixture} of shallow interpretable models using human-understandable concepts. As each interpretable model covers a subset of data, a mixture of interpretable models achieves comparable performance as BB. Further, we use the pseudo-labeling technique from semi-supervised learning (SSL) to learn the concept classifier in the target domain, followed by fine-tuning the interpretable models in the target domain. We evaluate our model using a real-life large-scale chest-X-ray (CXR) classification dataset. The code is available at: \url{https://github.com/batmanlab/MICCAI-2023-Route-interpret-repeat-CXRs}.
DR-VIDAL -- Doubly Robust Variational Information-theoretic Deep Adversarial Learning for Counterfactual Prediction and Treatment Effect Estimation on Real World Data
Mar 07, 2023Abstract:Determining causal effects of interventions onto outcomes from real-world, observational (non-randomized) data, e.g., treatment repurposing using electronic health records, is challenging due to underlying bias. Causal deep learning has improved over traditional techniques for estimating individualized treatment effects (ITE). We present the Doubly Robust Variational Information-theoretic Deep Adversarial Learning (DR-VIDAL), a novel generative framework that combines two joint models of treatment and outcome, ensuring an unbiased ITE estimation even when one of the two is misspecified. DR-VIDAL integrates: (i) a variational autoencoder (VAE) to factorize confounders into latent variables according to causal assumptions; (ii) an information-theoretic generative adversarial network (Info-GAN) to generate counterfactuals; (iii) a doubly robust block incorporating treatment propensities for outcome predictions. On synthetic and real-world datasets (Infant Health and Development Program, Twin Birth Registry, and National Supported Work Program), DR-VIDAL achieves better performance than other non-generative and generative methods. In conclusion, DR-VIDAL uniquely fuses causal assumptions, VAE, Info-GAN, and doubly robustness into a comprehensive, performant framework. Code is available at: https://github.com/Shantanu48114860/DR-VIDAL-AMIA-22 under MIT license.
Route, Interpret, Repeat: Blurring the Line Between Post hoc Explainability and Interpretable Models
Feb 20, 2023Abstract:The current approach to ML model design is either to choose a flexible Blackbox model and explain it post hoc or to start with an interpretable model. Blackbox models are flexible but difficult to explain, whereas interpretable models are designed to be explainable. However, developing interpretable models necessitates extensive ML knowledge, and the resulting models tend to be less flexible, offering potentially subpar performance compared to their Blackbox equivalents. This paper aims to blur the distinction between a post hoc explanation of a BlackBox and constructing interpretable models. We propose beginning with a flexible BlackBox model and gradually \emph{carving out} a mixture of interpretable models and a \emph{residual network}. Our design identifies a subset of samples and \emph{routes} them through the interpretable models. The remaining samples are routed through a flexible residual network. We adopt First Order Logic (FOL) as the interpretable model's backbone, which provides basic reasoning on concepts retrieved from the BlackBox model. On the residual network, we repeat the method until the proportion of data explained by the residual network falls below a desired threshold. Our approach offers several advantages. First, the mixture of interpretable and flexible residual networks results in almost no compromise in performance. Second, the route, interpret, and repeat approach yields a highly flexible interpretable model. Our extensive experiment demonstrates the performance of the model on various datasets. We show that by editing the FOL model, we can fix the shortcut learned by the original BlackBox model. Finally, our method provides a framework for a hybrid symbolic-connectionist network that is simple to train and adaptable to many applications.
Anatomy-Guided Weakly-Supervised Abnormality Localization in Chest X-rays
Jun 25, 2022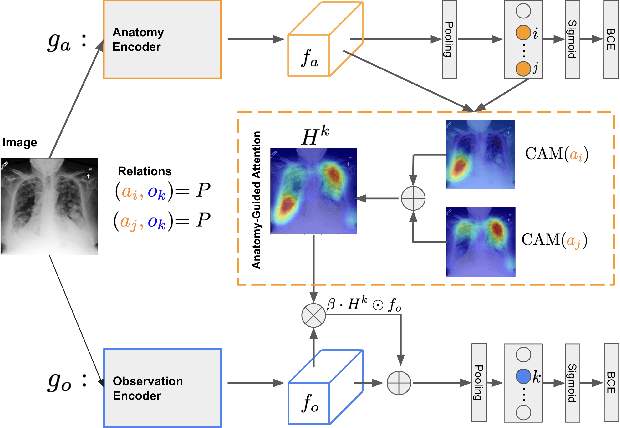
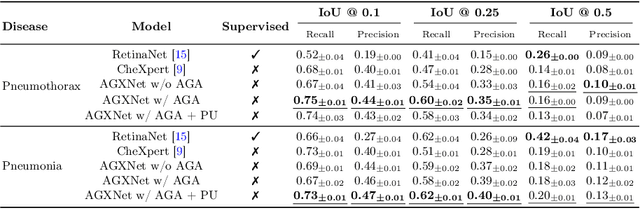
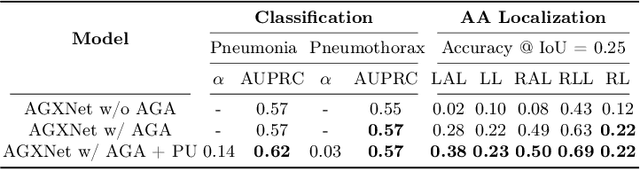
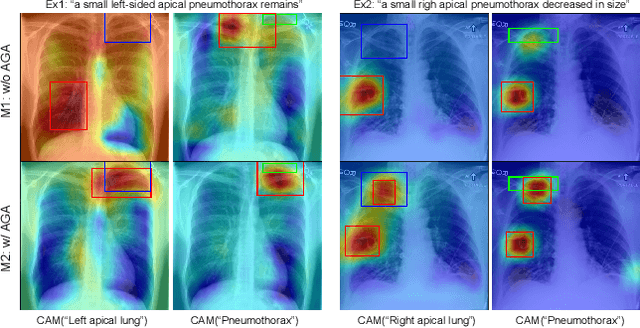
Abstract:Creating a large-scale dataset of abnormality annotation on medical images is a labor-intensive and costly task. Leveraging weak supervision from readily available data such as radiology reports can compensate lack of large-scale data for anomaly detection methods. However, most of the current methods only use image-level pathological observations, failing to utilize the relevant anatomy mentions in reports. Furthermore, Natural Language Processing (NLP)-mined weak labels are noisy due to label sparsity and linguistic ambiguity. We propose an Anatomy-Guided chest X-ray Network (AGXNet) to address these issues of weak annotation. Our framework consists of a cascade of two networks, one responsible for identifying anatomical abnormalities and the second responsible for pathological observations. The critical component in our framework is an anatomy-guided attention module that aids the downstream observation network in focusing on the relevant anatomical regions generated by the anatomy network. We use Positive Unlabeled (PU) learning to account for the fact that lack of mention does not necessarily mean a negative label. Our quantitative and qualitative results on the MIMIC-CXR dataset demonstrate the effectiveness of AGXNet in disease and anatomical abnormality localization. Experiments on the NIH Chest X-ray dataset show that the learned feature representations are transferable and can achieve the state-of-the-art performances in disease classification and competitive disease localization results. Our code is available at https://github.com/batmanlab/AGXNet
High Accuracy Classification of Parkinson's Disease through Shape Analysis and Surface Fitting in $^{123}$I-Ioflupane SPECT Imaging
Mar 04, 2017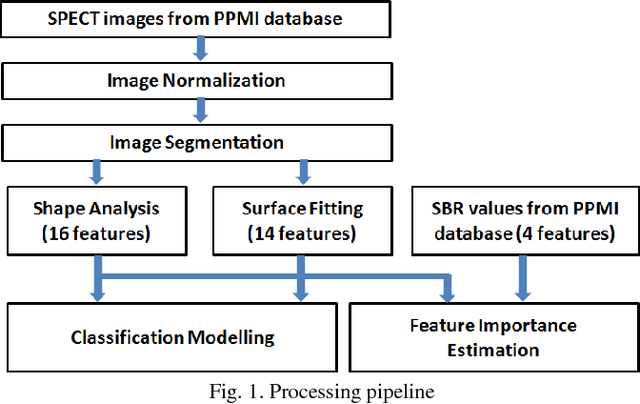
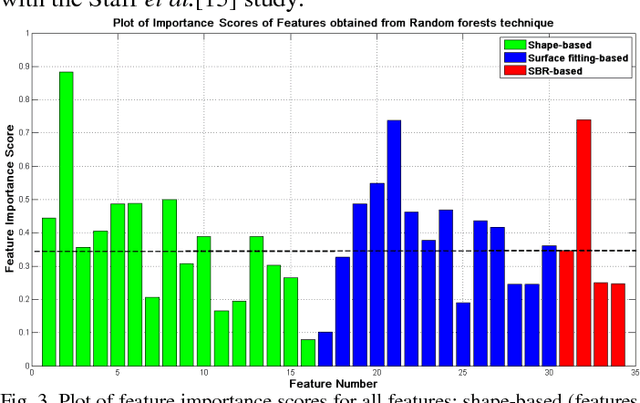
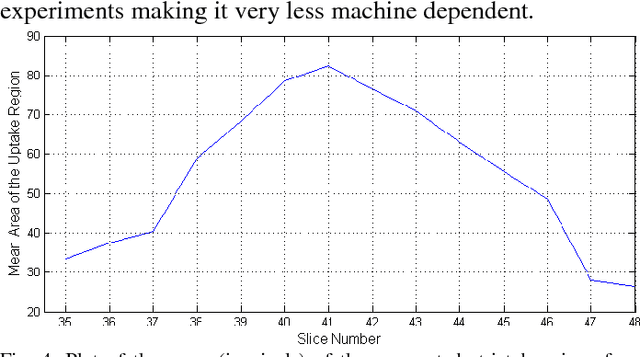
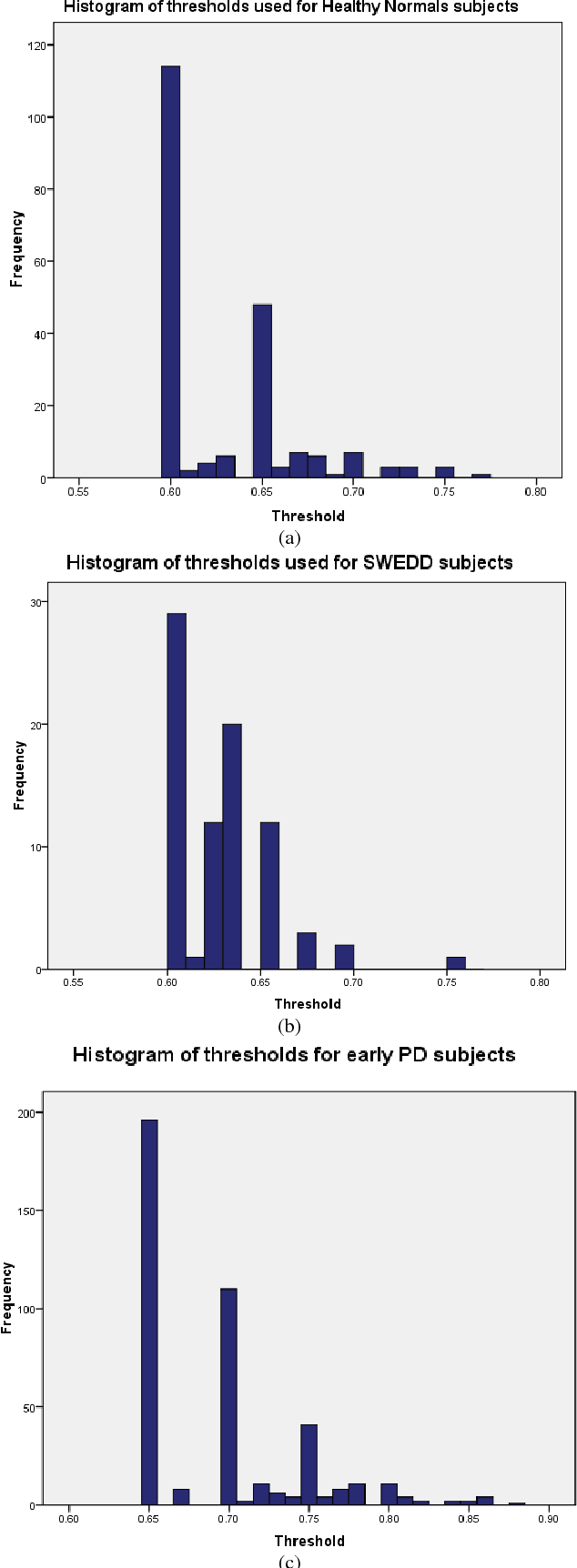
Abstract:Early and accurate identification of parkinsonian syndromes (PS) involving presynaptic degeneration from non-degenerative variants such as Scans Without Evidence of Dopaminergic Deficit (SWEDD) and tremor disorders, is important for effective patient management as the course, therapy and prognosis differ substantially between the two groups. In this study, we use Single Photon Emission Computed Tomography (SPECT) images from healthy normal, early PD and SWEDD subjects, as obtained from the Parkinson's Progression Markers Initiative (PPMI) database, and process them to compute shape- and surface fitting-based features for the three groups. We use these features to develop and compare various classification models that can discriminate between scans showing dopaminergic deficit, as in PD, from scans without the deficit, as in healthy normal or SWEDD. Along with it, we also compare these features with Striatal Binding Ratio (SBR)-based features, which are well-established and clinically used, by computing a feature importance score using Random forests technique. We observe that the Support Vector Machine (SVM) classifier gave the best performance with an accuracy of 97.29%. These features also showed higher importance than the SBR-based features. We infer from the study that shape analysis and surface fitting are useful and promising methods for extracting discriminatory features that can be used to develop diagnostic models that might have the potential to help clinicians in the diagnostic process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge