Zeqiang Wang
OrthoGeoLoRA: Geometric Parameter-Efficient Fine-Tuning for Structured Social Science Concept Retrieval on theWeb
Jan 14, 2026Abstract:Large language models and text encoders increasingly power web-based information systems in the social sciences, including digital libraries, data catalogues, and search interfaces used by researchers, policymakers, and civil society. Full fine-tuning is often computationally and energy intensive, which can be prohibitive for smaller institutions and non-profit organizations in the Web4Good ecosystem. Parameter-Efficient Fine-Tuning (PEFT), especially Low-Rank Adaptation (LoRA), reduces this cost by updating only a small number of parameters. We show that the standard LoRA update $ΔW = BA^\top$ has geometric drawbacks: gauge freedom, scale ambiguity, and a tendency toward rank collapse. We introduce OrthoGeoLoRA, which enforces an SVD-like form $ΔW = BΣA^\top$ by constraining the low-rank factors to be orthogonal (Stiefel manifold). A geometric reparameterization implements this constraint while remaining compatible with standard optimizers such as Adam and existing fine-tuning pipelines. We also propose a benchmark for hierarchical concept retrieval over the European Language Social Science Thesaurus (ELSST), widely used to organize social science resources in digital repositories. Experiments with a multilingual sentence encoder show that OrthoGeoLoRA outperforms standard LoRA and several strong PEFT variants on ranking metrics under the same low-rank budget, offering a more compute- and parameter-efficient path to adapt foundation models in resource-constrained settings.
Are Information Retrieval Approaches Good at Harmonising Longitudinal Survey Questions in Social Science?
Apr 29, 2025Abstract:Automated detection of semantically equivalent questions in longitudinal social science surveys is crucial for long-term studies informing empirical research in the social, economic, and health sciences. Retrieving equivalent questions faces dual challenges: inconsistent representation of theoretical constructs (i.e. concept/sub-concept) across studies as well as between question and response options, and the evolution of vocabulary and structure in longitudinal text. To address these challenges, our multi-disciplinary collaboration of computer scientists and survey specialists presents a new information retrieval (IR) task of identifying concept (e.g. Housing, Job, etc.) equivalence across question and response options to harmonise longitudinal population studies. This paper investigates multiple unsupervised approaches on a survey dataset spanning 1946-2020, including probabilistic models, linear probing of language models, and pre-trained neural networks specialised for IR. We show that IR-specialised neural models achieve the highest overall performance with other approaches performing comparably. Additionally, the re-ranking of the probabilistic model's results with neural models only introduces modest improvements of 0.07 at most in F1-score. Qualitative post-hoc evaluation by survey specialists shows that models generally have a low sensitivity to questions with high lexical overlap, particularly in cases where sub-concepts are mismatched. Altogether, our analysis serves to further research on harmonising longitudinal studies in social science.
Revealing COVID-19's Social Dynamics: Diachronic Semantic Analysis of Vaccine and Symptom Discourse on Twitter
Oct 10, 2024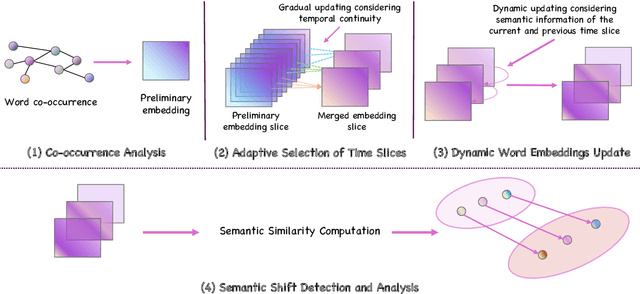
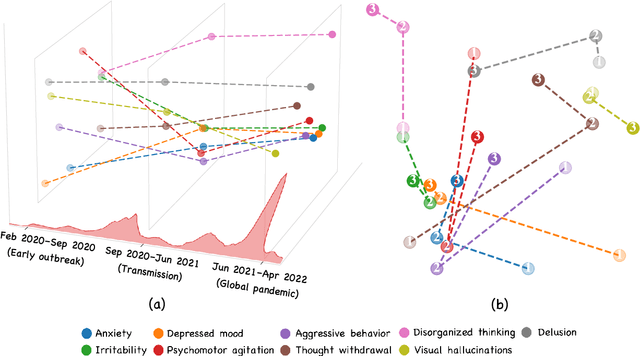
Abstract:Social media is recognized as an important source for deriving insights into public opinion dynamics and social impacts due to the vast textual data generated daily and the 'unconstrained' behavior of people interacting on these platforms. However, such analyses prove challenging due to the semantic shift phenomenon, where word meanings evolve over time. This paper proposes an unsupervised dynamic word embedding method to capture longitudinal semantic shifts in social media data without predefined anchor words. The method leverages word co-occurrence statistics and dynamic updating to adapt embeddings over time, addressing the challenges of data sparseness, imbalanced distributions, and synergistic semantic effects. Evaluated on a large COVID-19 Twitter dataset, the method reveals semantic evolution patterns of vaccine- and symptom-related entities across different pandemic stages, and their potential correlations with real-world statistics. Our key contributions include the dynamic embedding technique, empirical analysis of COVID-19 semantic shifts, and discussions on enhancing semantic shift modeling for computational social science research. This study enables capturing longitudinal semantic dynamics on social media to understand public discourse and collective phenomena.
DKE-Research at SemEval-2024 Task 2: Incorporating Data Augmentation with Generative Models and Biomedical Knowledge to Enhance Inference Robustness
Apr 14, 2024



Abstract:Safe and reliable natural language inference is critical for extracting insights from clinical trial reports but poses challenges due to biases in large pre-trained language models. This paper presents a novel data augmentation technique to improve model robustness for biomedical natural language inference in clinical trials. By generating synthetic examples through semantic perturbations and domain-specific vocabulary replacement and adding a new task for numerical and quantitative reasoning, we introduce greater diversity and reduce shortcut learning. Our approach, combined with multi-task learning and the DeBERTa architecture, achieved significant performance gains on the NLI4CT 2024 benchmark compared to the original language models. Ablation studies validate the contribution of each augmentation method in improving robustness. Our best-performing model ranked 12th in terms of faithfulness and 8th in terms of consistency, respectively, out of the 32 participants.
Zero-Shot Medical Information Retrieval via Knowledge Graph Embedding
Oct 31, 2023Abstract:In the era of the Internet of Things (IoT), the retrieval of relevant medical information has become essential for efficient clinical decision-making. This paper introduces MedFusionRank, a novel approach to zero-shot medical information retrieval (MIR) that combines the strengths of pre-trained language models and statistical methods while addressing their limitations. The proposed approach leverages a pre-trained BERT-style model to extract compact yet informative keywords. These keywords are then enriched with domain knowledge by linking them to conceptual entities within a medical knowledge graph. Experimental evaluations on medical datasets demonstrate MedFusion Rank's superior performance over existing methods, with promising results with a variety of evaluation metrics. MedFusionRank demonstrates efficacy in retrieving relevant information, even from short or single-term queries.
YATO: Yet Another deep learning based Text analysis Open toolkit
Sep 28, 2022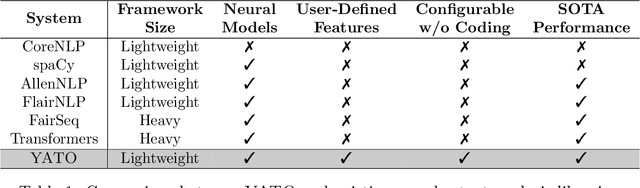

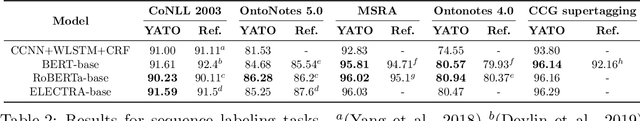

Abstract:We introduce YATO, an open-source toolkit for text analysis with deep learning. It focuses on fundamental sequence labeling and sequence classification tasks on text. Designed in a hierarchical structure, YATO supports free combinations of three types of features including 1) traditional neural networks (CNN, RNN, etc.); 2) pre-trained language models (BERT, RoBERTa, ELECTRA, etc.); and 3) user-customed neural features via a simple configurable file. Benefiting from the advantages of flexibility and ease of use, YATO can facilitate reproducing and refinement of state-of-the-art NLP models, and promote the cross-disciplinary applications of NLP techniques. Source code, examples, and documentation are publicly available at https://github.com/jiesutd/YATO.
METS-CoV: A Dataset of Medical Entity and Targeted Sentiment on COVID-19 Related Tweets
Sep 28, 2022



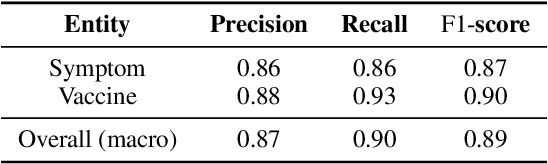
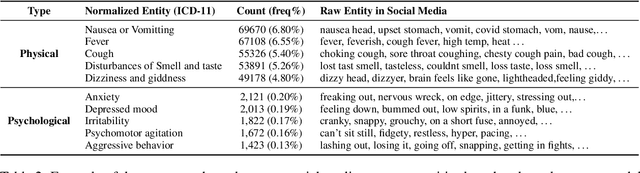
Abstract:The COVID-19 pandemic continues to bring up various topics discussed or debated on social media. In order to explore the impact of pandemics on people's lives, it is crucial to understand the public's concerns and attitudes towards pandemic-related entities (e.g., drugs, vaccines) on social media. However, models trained on existing named entity recognition (NER) or targeted sentiment analysis (TSA) datasets have limited ability to understand COVID-19-related social media texts because these datasets are not designed or annotated from a medical perspective. This paper releases METS-CoV, a dataset containing medical entities and targeted sentiments from COVID-19-related tweets. METS-CoV contains 10,000 tweets with 7 types of entities, including 4 medical entity types (Disease, Drug, Symptom, and Vaccine) and 3 general entity types (Person, Location, and Organization). To further investigate tweet users' attitudes toward specific entities, 4 types of entities (Person, Organization, Drug, and Vaccine) are selected and annotated with user sentiments, resulting in a targeted sentiment dataset with 9,101 entities (in 5,278 tweets). To the best of our knowledge, METS-CoV is the first dataset to collect medical entities and corresponding sentiments of COVID-19-related tweets. We benchmark the performance of classical machine learning models and state-of-the-art deep learning models on NER and TSA tasks with extensive experiments. Results show that the dataset has vast room for improvement for both NER and TSA tasks. METS-CoV is an important resource for developing better medical social media tools and facilitating computational social science research, especially in epidemiology. Our data, annotation guidelines, benchmark models, and source code are publicly available (https://github.com/YLab-Open/METS-CoV) to ensure reproducibility.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge